8KG2
 
 | |
8J07
 
 | |
7CTG
 
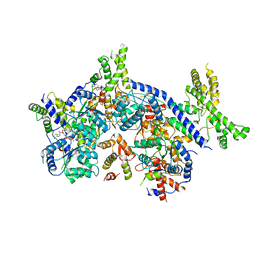 | | Human Origin Recognition Complex, ORC1-5 State I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7D4I
 
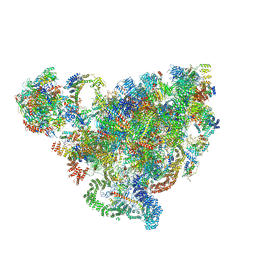 | | Cryo-EM structure of 90S small ribosomal precursors complex with the DEAH-box RNA helicase Dhr1 (State F) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of 90S small ribosomal precursors complex with Dhr1
To Be Published
|
|
7D63
 
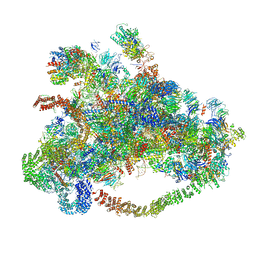 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D5S
 
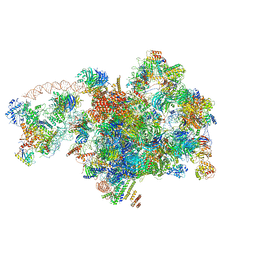 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
8OOI
 
 | | Full composite cryo-EM map of p97/VCP in ADP.Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheng, T.C, Sakata, E, Schuetz, A.K. | | Deposit date: | 2023-04-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Characterizing ATP processing by the AAA+ protein p97 at the atomic level.
Nat.Chem., 16, 2024
|
|
8OJL
 
 | | Human Mitochondrial Lon Y394E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OM7
 
 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OKA
 
 | | Human Mitochondrial Lon Y394F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OVF
 
 | | Human Mitochondrial Lon Y186F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.23 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OVG
 
 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.47 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
7FD4
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FID
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIZ
 
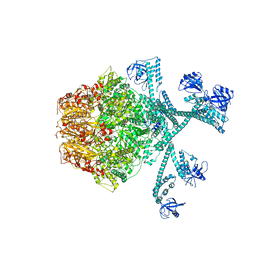 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-08-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIE
 
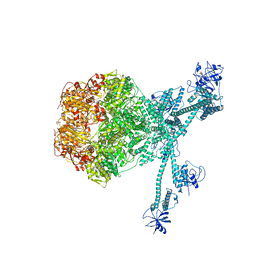 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
5WC0
 
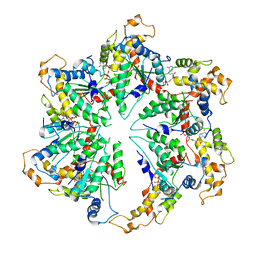 | | katanin hexamer in spiral conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Meiotic spindle formation protein mei-1 | | Authors: | Zehr, E.A, Szyk, A, Piszczek, G, Szczesna, E, Zuo, X, Roll-Mecak, A. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5WC1
 
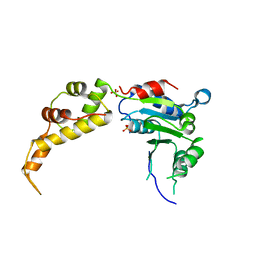 | | katanin AAA ATPase domain | | Descriptor: | Meiotic spindle formation protein mei-1, SULFATE ION | | Authors: | Szyk, A, Roll-Mecak, A. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5WBW
 
 | | Yeast Hsp104 fragment 1-360 | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase.
Biosci. Rep., 37, 2017
|
|
5WCB
 
 | | Katanin hexamer in the ring conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Meiotic spindle formation protein mei-1 | | Authors: | Zehr, E.A, Szyk, A, Piszczek, G, Szczesna, E, Zuo, X, Roll-Mecak, A. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5WYJ
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WYK
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2017-05-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
