9FH9
 
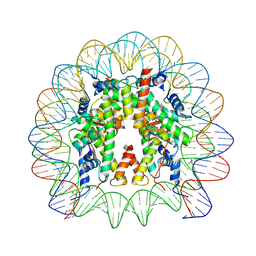 | | Structure of CyclinB1 N-terminus bound to the NCP | | Descriptor: | DNA (145-MER), G2/mitotic-specific cyclin-B1, Histone H2A type 3, ... | | Authors: | Young, R.V.C, Muhammad, R, Alfieri, C. | | Deposit date: | 2024-05-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
9FGQ
 
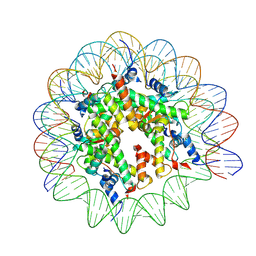 | | Structure of human APC3loop 375-381 bound to the NCP | | Descriptor: | Cell division cycle protein 27 homolog, DNA (131-MER), DNA (132-MER), ... | | Authors: | Young, R.V.C, Muhammad, R, Alfieri, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
4CDV
 
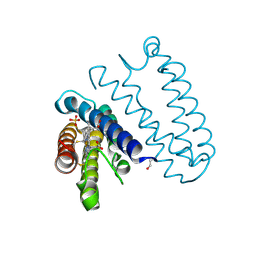 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 100K | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1GM4
 
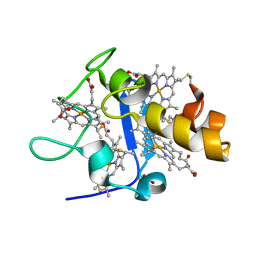 | | OXIDISED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-10 | | Release date: | 2002-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome.
J.Biol.Chem., 276, 2001
|
|
1GMB
 
 | | Reduced structure of CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome
J.Biol.Chem., 276, 2001
|
|
2QW6
 
 | | Crystal structure of the C-terminal domain of an AAA ATPase from Enterococcus faecium DO | | Descriptor: | AAA ATPase, central region | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of an AAA ATPase from Enterococcus faecium DO.
To be Published
|
|
3BGE
 
 | | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae | | Descriptor: | Predicted ATPase, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Meyer, A.J, Toro, R, Freeman, J, Adams, J, Koss, J, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae.
To be Published
|
|
2K3V
 
 | | Solution Structure of a Tetrahaem Cytochrome from Shewanella Frigidimarina | | Descriptor: | HEME C, Tetraheme cytochrome c-type | | Authors: | Paixao, V.B, Turner, D.L, Salgueiro, C.A, Brennan, L, Reid, G.A, Chapman, S.K. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a tetraheme cytochrome from Shewanella frigidimarina reveals a novel family structural motif
Biochemistry, 47, 2008
|
|
5CGD
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile - (HTL14242) | | Descriptor: | 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, OLEIC ACID | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
1FI3
 
 | |
7EAD
 
 | | Crystal structure of beta-sheet cytochrome c prime from Thermus thermophilus. | | Descriptor: | Cytochrome_P460 domain-containing protein, HEME C | | Authors: | Yoshimi, T, Fujii, S, Oki, H, Igawa, T, Adams, R.H, Ueda, K, Kawahara, K, Ohkubo, T, Hough, A.M, Sambongi, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of thermally stable homodimeric cytochrome c'-beta from Thermus thermophilus.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5CGC
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
5PZU
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor [5-[2-amino-5-(2-methylpropyl)-1,3-thiazol-4-yl]furan-2-yl]phosphonic acid | | Descriptor: | Fructose-1,6-bisphosphatase 1, {5-[2-amino-5-(2-methylpropyl)-1,3-thiazol-4-yl]furan-2-yl}phosphonic acid | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor [5-[2-amino-5-(2-methylpropyl)-1,3-thiazol-4-yl]furan-2-yl]phosphonic acid
To be published
|
|
5XK1
 
 | | Structure of 8-mer DNA3 | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*CP*CP*CP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3IGN
 
 | | Crystal Structure of the GGDEF domain from Marinobacter aquaeolei diguanylate cyclase complexed with c-di-GMP - Northeast Structural Genomics Consortium Target MqR89a | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate cyclase | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product.
J.Struct.Funct.Genom., 13, 2012
|
|
5Q02
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(1,2-oxazol-3-yl)thiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(1,2-oxazol-3-yl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(1,2-oxazol-3-yl)thiophen-2-yl]sulfonylurea
To be published
|
|
5Q0C
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with fructose-2,6-diphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(4-methoxyphenyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with f2,6p
To be published
|
|
4KYN
 
 | |
2CCP
 
 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
2CXB
 
 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-04-21 | | Release date: | 1995-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5B6O
 
 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2L18
 
 | |
4CJO
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced at 180K using X-rays | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CG4
 
 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
4L53
 
 | | Crystal Structure of (1R,4R)-4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)-3-chlorofuro[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}cyclohexan-1-ol bound to TAK1-TAB1 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, ... | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1: Optimization of kinase selectivity and pharmacokinetics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
