1TIM
 
 | | STRUCTURE OF TRIOSE PHOSPHATE ISOMERASE FROM CHICKEN MUSCLE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Banner, D.W, Bloomer, A.C, Petsko, G.A, Phillips, D.C, Wilson, I.A. | | Deposit date: | 1976-09-01 | | Release date: | 1976-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for triose phosphate isomerase from chicken muscle.
Biochem.Biophys.Res.Commun., 72, 1976
|
|
1STP
 
 | |
1TME
 
 | | THREE-DIMENSIONAL STRUCTURE OF THEILER VIRUS | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1992-01-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of Theiler virus.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1TDA
 
 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-02-15 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
2BWU
 
 | | Asp271Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
1THY
 
 | |
2BUE
 
 | | Structure of AAC(6')-Ib in complex with Ribostamycin and Coenzyme A. | | Descriptor: | AAC(6')-IB, CALCIUM ION, COENZYME A, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
2BWX
 
 | | His354Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BI8
 
 | |
2C1F
 
 | |
1STR
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STS
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
2C0R
 
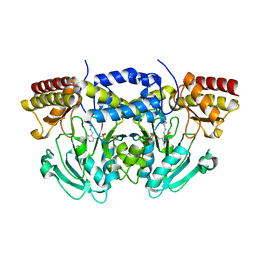 | |
1TOK
 
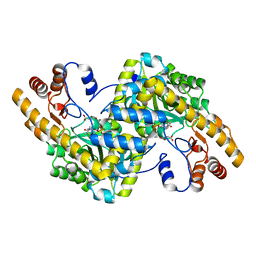 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
2BQ1
 
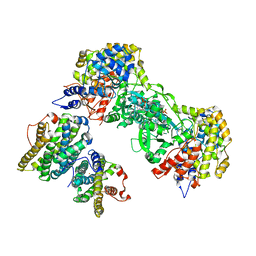 | | Ribonucleotide reductase class 1b holocomplex R1E,R2F from Salmonella typhimurium | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Uppsten, M, Farnegardh, M, Domkin, V, Uhlin, U. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | The First Holocomplex Structure of Ribonucleotide Reductase Gives New Insight Into its Mechanism of Action
J.Mol.Biol., 359, 2006
|
|
1TN5
 
 | | Structure of bacterorhodopsin mutant K41P | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Yang, D, Faham, S, Boulting, G, Whitelegge, J, Bowie, J.U. | | Deposit date: | 2004-06-11 | | Release date: | 2004-10-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline substitutions are not easily accommodated in a membrane protein
J.Mol.Biol., 341, 2004
|
|
1TO1
 
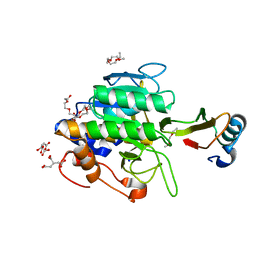 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 Y61A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
2BWV
 
 | | His361Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
1TN7
 
 | | Protein Farnesyltransferase Complexed with a TC21 Peptide Substrate and a FPP Analog at 2.3A Resolution | | Descriptor: | ACETIC ACID, Fusion protein, Protein farnesyltransferase alpha subunit, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
2BWW
 
 | | His350Ala Escherichia coli Aminopeptidase P | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AMINOPEPTIDASE P, CITRATE ANION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
1Z8N
 
 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With An Imidazolinone Herbicide, Imazaquin | | Descriptor: | 2-(4-ISOPROPYL-4-METHYL-5-OXO-4,5-DIHYDRO-1H-IMIDAZOL-2-YL)QUINOLINE-3-CARBOXYLIC ACID, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-03-31 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1X29
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-2-methyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-2-METHYL-L-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
1Z8Y
 
 | | Mapping the E2 Glycoprotein of Alphaviruses | | Descriptor: | Capsid protein C, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Mukhopadhyay, S, Zhang, W, Gabler, S, Chipman, P.R, Strauss, E.G, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-03-31 | | Release date: | 2006-02-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses.
Structure, 14, 2006
|
|
1ZJ3
 
 | | Crystal Structure of Human Galactosyltransferase (GTB) Complexed with H type II Trisaccharide | | Descriptor: | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-28 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
1WMV
 
 | |
