1PKZ
 
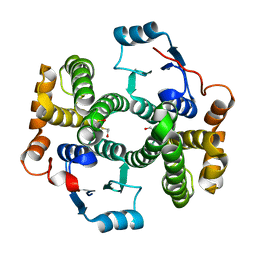 | | Crystal structure of human glutathione transferase (GST) A1-1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4F0B
 
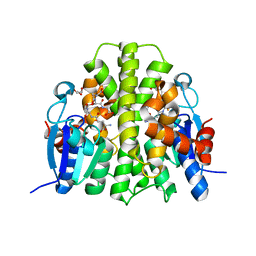 | |
4F0C
 
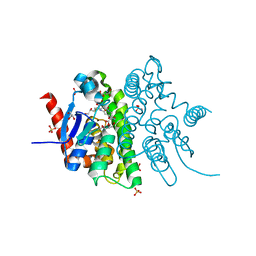 | | Crystal structure of the glutathione transferase URE2P5 from Phanerochaete chrysosporium | | Descriptor: | GLYCEROL, Glutathione transferase, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Didierjean, C, Favier, F, Roret, T. | | Deposit date: | 2012-05-04 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary divergence of Ure2pA glutathione transferases in wood degrading fungi.
Fungal Genet Biol, 83, 2015
|
|
3LG6
 
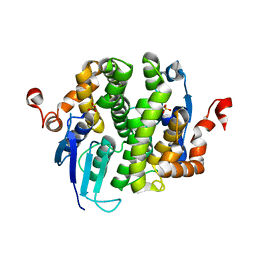 | |
7ZA5
 
 | | GSTF sh101 mutant | | Descriptor: | Glutathione transferase | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
3LSZ
 
 | | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides
To be Published
|
|
3LXZ
 
 | |
3LYK
 
 | |
3LYP
 
 | |
7ZZN
 
 | |
8A0I
 
 | |
8A0O
 
 | |
8A08
 
 | |
8A0R
 
 | |
8A0Q
 
 | |
8A0P
 
 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
3MAK
 
 | |
8ALS
 
 | |
3M3M
 
 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
3MDK
 
 | |
3N5O
 
 | |
7ZA4
 
 | | GSTF sh155 mutant | | Descriptor: | Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
8AGQ
 
 | | Crystal structure of anthocyanin-related GSTF8 from Populus trichocarpa in complex with (-)-catechin and glutathione | | Descriptor: | (2~{S},3~{R})-2-[3,4-bis(oxidanyl)phenyl]-3,4-dihydro-2~{H}-chromene-3,5,7-triol, GLUTATHIONE, Glutathione transferase, ... | | Authors: | Eichenberger, M, Hueppi, S, Schwander, T, Mittl, P, Buller, M.R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.093 Å) | | Cite: | The catalytic role of glutathione transferases in heterologous anthocyanin biosynthesis.
Nat Catal, 6, 2023
|
|
8BHZ
 
 | |
3M8N
 
 | |
