6PYJ
 
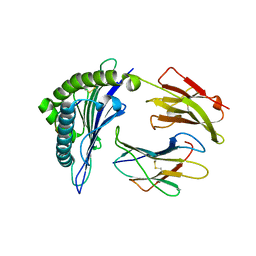 | | Crystal Structure of HLA-B*2705 in complex with LRN, a self-peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Gras, S. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Allelic association with ankylosing spondylitis fails to correlate with human leukocyte antigen B27 homodimer formation.
J.Biol.Chem., 294, 2019
|
|
3CJ3
 
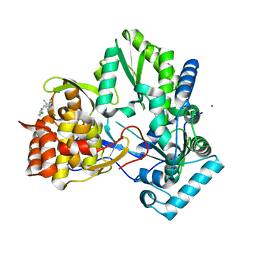 | |
3CJ2
 
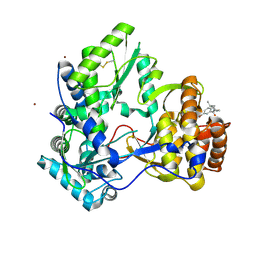 | |
3CND
 
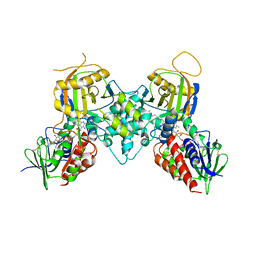 | | Crystal structure of fms1 in complex with N1-AcSpermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fms1 in complex with N1-AcSpermine
TO BE PUBLISHED
|
|
3CNT
 
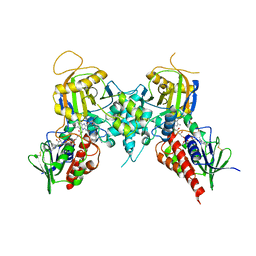 | |
3CN4
 
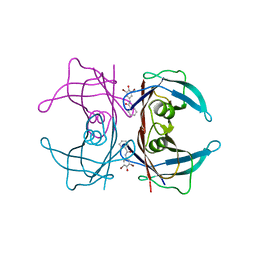 | |
3CNL
 
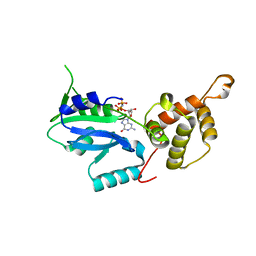 | | Crystal structure of GNP-bound YlqF from T. maritima | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3CNS
 
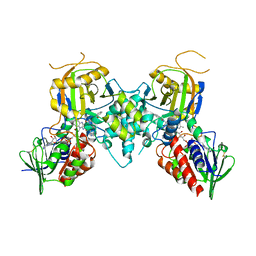 | |
3MM4
 
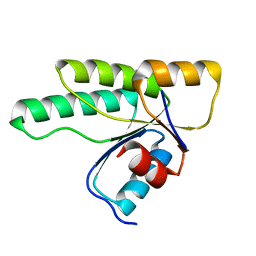 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase homolog | | Authors: | Marek, J, Klumpler, T, Pekarova, B, Triskova, O, Horak, J, Zidek, L, Dopitova, R, Hejatko, J, Janda, L. | | Deposit date: | 2010-04-19 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana.
Plant J., 67, 2011
|
|
6Q8E
 
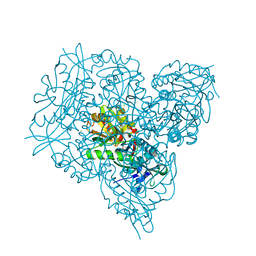 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain-amino-acid aminotransferase, CHLORIDE ION | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6PWI
 
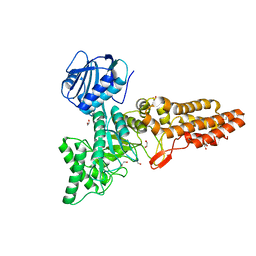 | | Structure of CpGH84D | | Descriptor: | 1,2-ETHANEDIOL, Putative hyaluronoglucosaminidase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
6PWT
 
 | |
3CI4
 
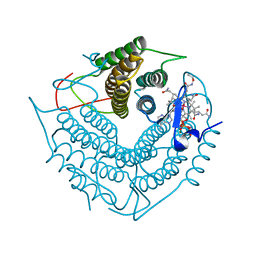 | | Structure of the PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri complexed with four-coordinate cob(II)inamide and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COB(II)INAMIDE, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | Maurice, M.St, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a human-type corrinoid adenosyltransferase confirms that coenzyme B12 is synthesized through a four-coordinate intermediate.
Biochemistry, 47, 2008
|
|
6ZA7
 
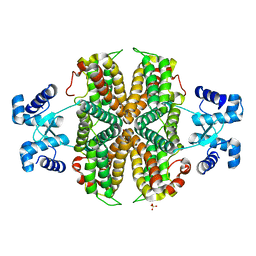 | | Structure of the apo transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
3MQ3
 
 | |
3D5F
 
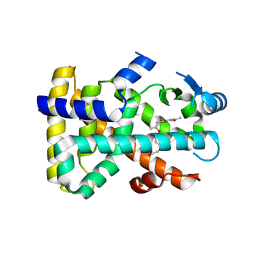 | | Crystal Structure of PPAR-delta complex | | Descriptor: | Peroxisome proliferator-activated receptor delta, {4-[3-(4-acetyl-3-hydroxy-2-propylphenoxy)propoxy]phenoxy}acetic acid | | Authors: | Amano, Y. | | Deposit date: | 2008-05-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PPAR-Delta Activation Contributes to Neuroprotectio Against Thapsigargin-Induced SH-SY5Y Cell Death
to be published
|
|
7Q34
 
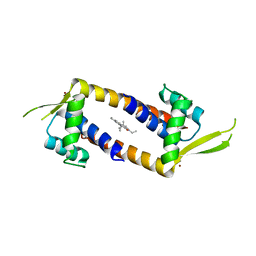 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in complex squaraine dye | | Descriptor: | 2,4-bis[(E)-(1-ethyl-3,3-dimethyl-indol-2-ylidene)methyl]cyclobutane-1,3-dione, Helix-turn-helix transcriptional regulator, NICKEL (II) ION | | Authors: | Liutkus, M, Mejias, S.H, Barolo, C, Cortajarena, A.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designing Artificial Fluorescent Proteins: Squaraine-LmrR Biophosphors for High Performance Deep-Red Biohybrid Light-Emitting Diodes
Adv Funct Mater, 32, 2022
|
|
3D62
 
 | | Development of Broad-Spectrum Halomethyl Ketone Inhibitors Against Coronavirus Main Protease 3CLpro | | Descriptor: | 3C-like proteinase, benzyl (2-oxopropyl)carbamate | | Authors: | Bacha, U, Barrila, J, Gabelli, S.B, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2008-05-18 | | Release date: | 2008-07-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of broad-spectrum halomethyl ketone inhibitors against coronavirus main protease 3CL(pro).
Chem.Biol.Drug Des., 72, 2008
|
|
6ZDY
 
 | | Crystal structure of WT murine S100A9 bound to calcium and zinc | | Descriptor: | CALCIUM ION, Protein S100-A9, SULFATE ION, ... | | Authors: | Yatime, L. | | Deposit date: | 2020-06-15 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Divalent cations influence the dimerization mode of murine S100A9 protein by modulating its disulfide bond pattern.
J.Struct.Biol., 213, 2020
|
|
6Q9U
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 12 AT 2.4A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-[(4~{S})-5-(5-chloranyl-2-oxidanylidene-1~{H}-pyridin-3-yl)-2-[2-(dimethylamino)-4-methoxy-pyrimidin-5-yl]-6-oxidanylidene-3-propan-2-yl-4~{H}-pyrrolo[3,4-c]pyrazol-4-yl]benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6PYW
 
 | | Crystal Structure of HLA-B*2705-W60A in complex with LRN, a self-peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Gras, S. | | Deposit date: | 2019-07-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Allelic association with ankylosing spondylitis fails to correlate with human leukocyte antigen B27 homodimer formation.
J.Biol.Chem., 294, 2019
|
|
3MBE
 
 | | TCR 21.30 in complex with MHC class II I-Ag7HEL(11-27) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II H2-IAg7 ALPHA CHAIN, ... | | Authors: | Corper, A.L, Yoshida, K, Teyton, L, Wilson, I.A. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | The diabetogenic mouse MHC class II molecule I-Ag7 is endowed with a switch that modulates TCR affinity.
J.Clin.Invest., 120, 2010
|
|
3MEW
 
 | | Crystal structure of Novel Tudor domain-containing protein SGF29 | | Descriptor: | SAGA-associated factor 29 homolog | | Authors: | Xu, C, Bian, C.B, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3CWE
 
 | | PTP1B in complex with a phosphonic acid inhibitor | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1, [{2-bromo-4-[(2R)-3-oxo-2,3-diphenylpropyl]phenyl}(difluoro)methyl]phosphonic acid | | Authors: | Scapin, G, Han, Y, Kennedy, B.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of [(3-bromo-7-cyano-2-naphthyl)(difluoro)methyl]phosphonic acid, a potent and orally active small molecule PTP1B inhibitor
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3ML0
 
 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in tetragonal form | | Descriptor: | CALCIUM ION, Penicillin G acylase, alpha subunit, ... | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
