3D04
 
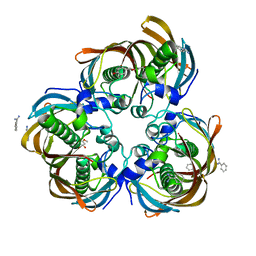 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with sakuranetin | | Descriptor: | (2S)-5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-2,3-dihydro-4H-chromen-4-one, (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, ... | | Authors: | Zhang, L, Kong, Y, Wu, D, Shen, X, Jiang, H. | | Deposit date: | 2008-05-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3DCZ
 
 | |
3DEE
 
 | |
3DB0
 
 | |
3DAO
 
 | |
3DLC
 
 | |
3DDD
 
 | |
3D82
 
 | |
3D8P
 
 | |
3D9R
 
 | |
3DB7
 
 | |
3DDJ
 
 | |
3DFU
 
 | |
3DI5
 
 | |
3DCF
 
 | |
4RU2
 
 | |
3DFE
 
 | |
3DKQ
 
 | |
3DL1
 
 | |
3DCX
 
 | |
3DI4
 
 | |
3DIN
 
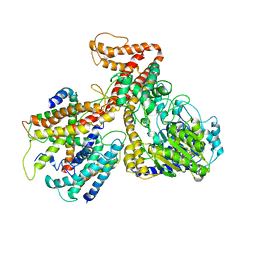 | | Crystal structure of the protein-translocation complex formed by the SecY channel and the SecA ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Zimmer, J, Nam, Y, Rapoport, T.A. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
3DJM
 
 | |
3DH4
 
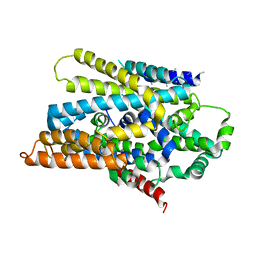 | | Crystal Structure of Sodium/Sugar symporter with bound Galactose from vibrio parahaemolyticus | | Descriptor: | ERBIUM (III) ION, SODIUM ION, Sodium/glucose cotransporter, ... | | Authors: | Abramson, J, Faham, S, Cascio, D. | | Deposit date: | 2008-06-16 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport.
Science, 321, 2008
|
|
3DO6
 
 | |
