6F7E
 
 | |
1M6A
 
 | |
5Z55
 
 | |
6B9W
 
 | |
6BB6
 
 | |
6BAM
 
 | |
1N6U
 
 | | NMR structure of the interferon-binding ectodomain of the human interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
1PBU
 
 | | Solution structure of the C-terminal domain of the human eEF1Bgamma subunit | | Descriptor: | Elongation factor 1-gamma | | Authors: | Vanwetswinkel, S, Kriek, J, Andersen, G.R, Guntert, P, Dijk, J, Canters, G.W, Siegal, G. | | Deposit date: | 2003-05-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, (15)N and (13)C resonance assignments of the highly conserved 19 kDa C-terminal domain from human Elongation Factor 1Bgamma.
J.Biomol.Nmr, 26, 2003
|
|
1C8P
 
 | | NMR STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE COMMON BETA-CHAIN IN THE GM-CSF, IL-3 AND IL-5 RECEPTORS | | Descriptor: | CYTOKINE RECEPTOR COMMON BETA CHAIN | | Authors: | Mulhern, T.D, D'Andrea, R.J, Gaunt, C, Vandeleur, L, Vadas, M.A, Lopez, A.F, Booker, G.W, Bagley, C.J. | | Deposit date: | 1999-10-05 | | Release date: | 2000-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the cytokine-binding domain of the common beta-chain of the receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5.
J.Mol.Biol., 297, 2000
|
|
1QRJ
 
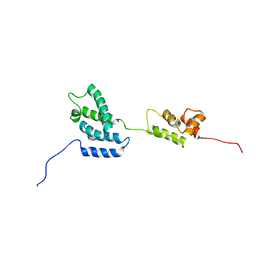 | | Solution structure of htlv-i capsid protein | | Descriptor: | HTLV-I CAPSID PROTEIN | | Authors: | Khorasanizadeh, S, Campos-Olivas, R, Clark, C.A, Summers, M.F. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H, 13C and 15N chemical shift assignment and secondary structure of the HTLV-I capsid protein.
J.Biomol.NMR, 14, 1999
|
|
1AZE
 
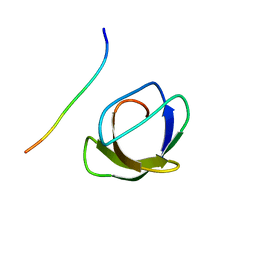 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE C32S-Y7V MUTANT OF THE NSH3 DOMAIN OF GRB2 WITH A PEPTIDE FROM SOS, 10 STRUCTURES | | Descriptor: | GRB2, SOS | | Authors: | Vidal, M, Gincel, E, Goudreau, N, Cornille, F, Parker, F, Duchesne, M, Tocque, B, Garbay, C, Roques, B.P. | | Deposit date: | 1997-11-17 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular and cellular analysis of Grb2 SH3 domain mutants: interaction with Sos and dynamin.
J.Mol.Biol., 290, 1999
|
|
1F70
 
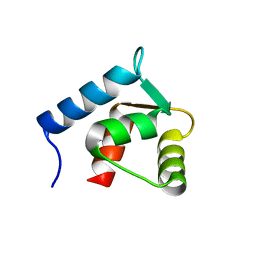 | |
1OVQ
 
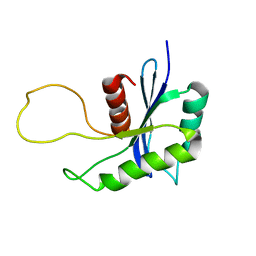 | |
1OZZ
 
 | | NMR structure of antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | defensin ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
1P00
 
 | | NMR structure of ETD151, mutant of the antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | defensin ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
1F71
 
 | |
6Z41
 
 | |
6Z40
 
 | |
1P0A
 
 | | NMR structure of ETD135, mutant of the antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | DEFENSIN ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
2WWV
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2WCY
 
 | | NMR solution structure of factor I-like modules of complement C7. | | Descriptor: | COMPLEMENT COMPONENT C7 | | Authors: | Phelan, M.M, Thai, C.T, Soares, D.C, Ogata, R.T, Barlow, P.N, Bramham, J. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Factor I-Like Modules from Complement C7 Reveals a Pair of Follistatin Domains in Compact Pseudosymmetric Arrangement.
J.Biol.Chem., 284, 2009
|
|
1FW7
 
 | |
2RPV
 
 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
7OD2
 
 | |
2WY2
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
