8ZAM
 
 | | EndoChR2 channelrhodopsin | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
8XHW
 
 | | Haloquadratum walsbyi middle rhodopsin mutant - D84N | | Descriptor: | Bacteriorhodopsin-II-like protein, RETINAL | | Authors: | Ko, L.N, Lim, G.Z, Li, G.Y, Chen, J.C, Yang, C.S. | | Deposit date: | 2023-12-18 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rhodopsin from Haloquadratum walsbyi is a light-driven magnesium transporter.
Nat Commun, 16, 2025
|
|
1P8I
 
 | | F219L BACTERIORHODOPSIN MUTANT | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Structures of the M and N Intermediates of Bacteriorhodopsin: Assembly of a Hydrogen-Bonded Chain of Water Molecules between Asp-96 and the Retinal Schiff Base
J.Mol.Biol., 330, 2003
|
|
1MGY
 
 | | Structure of the D85S mutant of bacteriorhodopsin with bromide bound | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, BROMIDE ION, Bacteriorhodopsin, ... | | Authors: | Facciotti, M.T, Cheung, V.S, Nguyen, D, Rouhani, S, Glaeser, R.M. | | Deposit date: | 2002-08-16 | | Release date: | 2003-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bromide-Bound D85S Mutant of Bacteriorhodopsin:
Principles of Ion Pumping
Biophys.J., 85, 2003
|
|
1P8U
 
 | | BACTERIORHODOPSIN N' INTERMEDIATE AT 1.62 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic Structures of the M and N Intermediates of Bacteriorhodopsin: Assembly of a Hydrogen-Bonded Chain of Water Molecules between Asp-96 and the Retinal Schiff Base
J.Mol.Biol., 330, 2003
|
|
1P8H
 
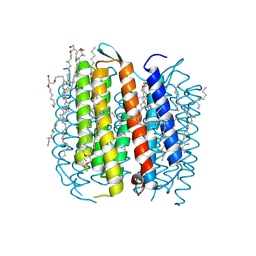 | | BACTERIORHODOPSIN M1 INTERMEDIATE PRODUCED AT ROOM TEMPERATURE | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic Structures of the M and N Intermediates of Bacteriorhodopsin: Assembly of a Hydrogen-Bonded Chain of Water Molecules between Asp-96 and the Retinal Schiff Base
J.Mol.Biol., 330, 2003
|
|
8PWG
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 90-120 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Ortolani, G, Bosman, R, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWI
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 60-90 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWQ
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 120-150 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWJ
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography. 30-60 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWP
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 0-30 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Ortolani, G, Bosman, R, Branden, G, Neutze, R. | | Deposit date: | 2023-07-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
4JR8
 
 | |
3UTY
 
 | |
8QLE
 
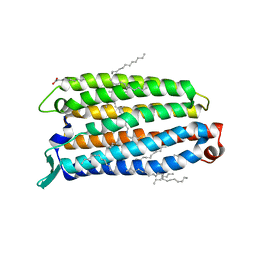 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLF
 
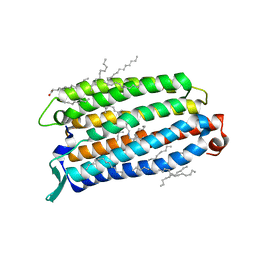 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QQZ
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
3UTV
 
 | | Crystal structure of bacteriorhodopsin mutant Y57F | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bacteriorhodopsin, RETINAL | | Authors: | Cao, Z, Bowie, J.U. | | Deposit date: | 2011-11-26 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Shifting hydrogen bonds may produce flexible transmembrane helices.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VHZ
 
 | |
3UTX
 
 | |
3UG9
 
 | | Crystal Structure of the Closed State of Channelrhodopsin | | Descriptor: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | Authors: | Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2011-11-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HYJ
 
 | | Crystal structure of Exiguobacterium sibiricum rhodopsin | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Gushchin, I, Chervakov, P, Kuzmichev, P, Popov, A, Round, E, Borshchevskiy, V, Dolgikh, D, Kirpichnikov, M, Petrovskaya, L, Chupin, V, Arseniev, A, Gordeliy, V. | | Deposit date: | 2012-11-13 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UTW
 
 | | Crystal structure of bacteriorhodopsin mutant P50A/Y57F | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bacteriorhodopsin, RETINAL, ... | | Authors: | Cao, Z, Bowie, J.U. | | Deposit date: | 2011-11-27 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shifting hydrogen bonds may produce flexible transmembrane helices.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
