6AIF
 
 | |
5WLP
 
 | |
6A42
 
 | | R1EN(5-223)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
8DQJ
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (AST) bound to ATP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
6L95
 
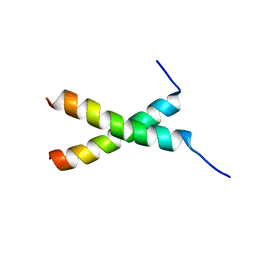 | | transmembrane-domain of Bax | | Descriptor: | Apoptosis regulator BAX | | Authors: | Bo, O.Y, Fujiao, L. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | transmembrane-domain of Bax
To Be Published
|
|
6ZXQ
 
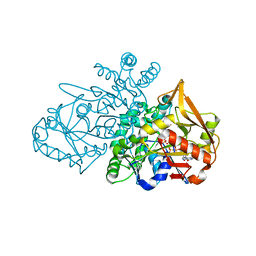 | | Adenylosuccinate Synthetase from H. pylori in complex with HDA, GDP, IMO, Mg | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bubic, A, Stefanic, Z. | | Deposit date: | 2020-07-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The pursuit of new alternative ways to eradicate Helicobacter pylori continues: Detailed characterization of interactions in the adenylosuccinate synthetase active site
Int.J.Biol.Macromol., 2023
|
|
6B7Q
 
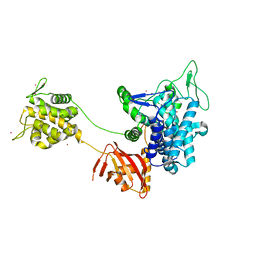 | |
6B7M
 
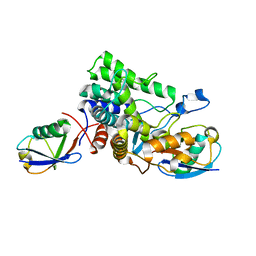 | |
7AG7
 
 | |
7AG4
 
 | |
7AG1
 
 | |
7AG6
 
 | |
7AGK
 
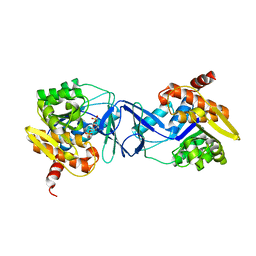 | | Crystal structure of E. coli SF kinase (YihV) in complex with product sulfofructose phosphate (SFP) | | Descriptor: | Sulfofructose kinase, [(2~{S},3~{S},4~{S},5~{R})-3,4,5-tris(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
6B7O
 
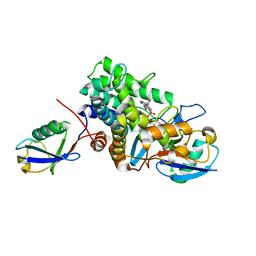 | | Crystal structure of Legionella effector sdeD (lpg2509) H67A in complex with ADP-ribosylated Ubiquitin | | Descriptor: | Polyubiquitin-C, SdeD (lpg2509) H67A, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Mao, Y, Akturk, A, Wasilko, J. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector.
Nature, 557, 2018
|
|
6B7P
 
 | |
6C9S
 
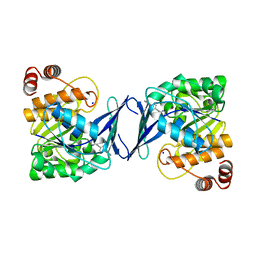 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3R,4S,5R)-2-(6-([1,1'-biphenyl]-4-ylethynyl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 6-[([1,1'-biphenyl]-4-yl)ethynyl]-9-beta-D-ribofuranosyl-9H-purine, Adenosine kinase, SODIUM ION, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
8EDD
 
 | | Staphylococcus aureus endonuclease IV Y33F mutant | | Descriptor: | CHLORIDE ION, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Saper, M.A, Kirillov, S, Isupov, M.N, Wiener, R, Rouvinski, A. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
To Be Published
|
|
7ALE
 
 | | Crystal structure of human PAICS in complex with inhibitor 69 | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 2-azanyl-~{N}-[2-bromanyl-5-[4-[3-(dimethylamino)propylsulfonyl]piperazin-1-yl]phenyl]-1,3-oxazole-4-carboxamide, Multifunctional protein ADE2 | | Authors: | Skerlova, J, Marttila, P, Unterlass, J, Jemth, A.-S, Henriksson, M, Wakchaure, P, Grube, M, Warpman Berglund, U, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cellular and biochemical validation of a potent PAICS inhibitor
To Be Published
|
|
6C9V
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | (2R,3S,4R,5R)-2-(hydroxymethyl)-5-[6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl]tetrahydrofuran-3,4-diol, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
7C0V
 
 | | Crystal structure of a dinucleotide-binding protein (Y224A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, CARBON DIOXIDE, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0L
 
 | |
7C0X
 
 | | Crystal structure of a dinucleotide-binding protein (T240A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CARBON DIOXIDE, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0K
 
 | | Crystal structure of a dinucleotide-binding protein of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-HYDROXY BUTANE-1,4-DIOL, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0Y
 
 | | Crystal structure of a dinucleotide-binding protein (Y246A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, GLYCEROL, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0O
 
 | | Crystal structure of a dinucleotide-binding protein (Y56F) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CARBONATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
