3NB8
 
 | | Crystal structure of oxidized H88Q Synechocystis sp. PCYA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Gae, D.D. | | Deposit date: | 2010-06-03 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
1MKK
 
 | | Disulfide deficient mutant of vascular endothelial growth factor A (C61A and C104A) | | Descriptor: | Vascular Endothelial Growth Factor A | | Authors: | Muller, Y.A, Heiring, C, Misselwitz, R, Welfle, K, Welfle, H. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The cystine knot promotes folding and not thermodynamic stability in vascular endothelial growth factor
J.Biol.Chem., 277, 2002
|
|
1MJV
 
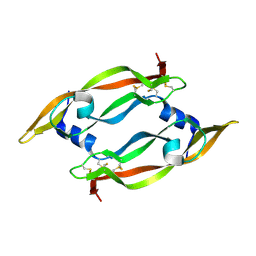 | | DISULFIDE DEFICIENT MUTANT OF VASCULAR ENDOTHELIAL GROWTH FACTOR A (C51A and C60A) | | Descriptor: | Vascular Endothelial Growth Factor A | | Authors: | Muller, Y.A, Heiring, C, Misselwitz, R, Welfle, K, Welfle, H. | | Deposit date: | 2002-08-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cystine knot promotes folding and not thermodynamic stability in vascular endothelial growth factor
J.Biol.Chem., 277, 2002
|
|
6BIG
 
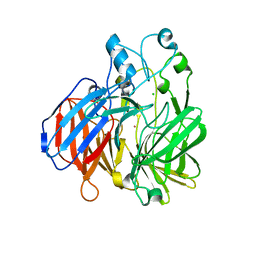 | | Crystal structure of cobalt-substituted Synechocystis ACO | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, COBALT (II) ION | | Authors: | Sui, X, Shi, W, Kiser, P.D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Preparation and characterization of metal-substituted carotenoid cleavage oxygenases.
J. Biol. Inorg. Chem., 23, 2018
|
|
6IR4
 
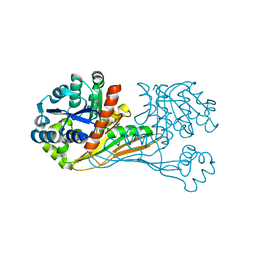 | | Crystal structure of BioU from Synechocystis sp.PCC6803 (apo form) | | Descriptor: | Slr0355 protein | | Authors: | Sakaki, K, Oishi, K, Shimizu, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
2QJO
 
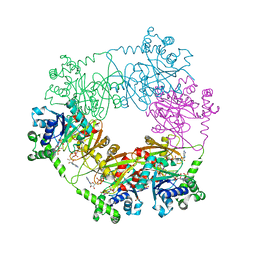 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
1NP7
 
 | | Crystal Structure Analysis of Synechocystis sp. PCC6803 cryptochrome | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Brudler, R, Hitomi, K, Daiyasu, H, Toh, H, Kucho, K, Ishiura, M, Kanehisa, M, Roberts, V.A, Todo, T, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new cryptochrome class: structure, function, and evolution
Mol.Cell, 11, 2003
|
|
6ITD
 
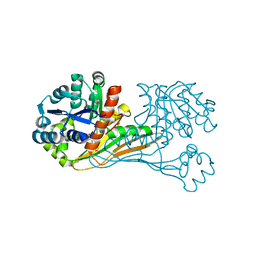 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-21 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6K38
 
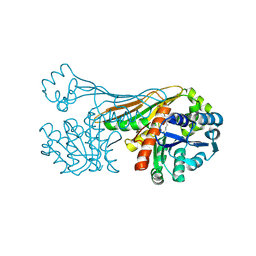 | |
6K37
 
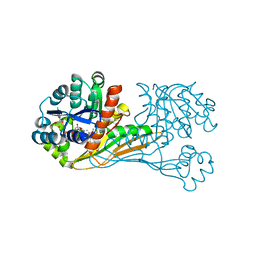 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with NAD+ and the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | (3R)-3-[(1R)-1-azanylethyl]nonanedioic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Nishiyama, M. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
4F0T
 
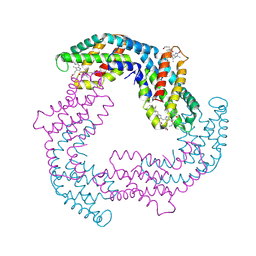 | |
2HA8
 
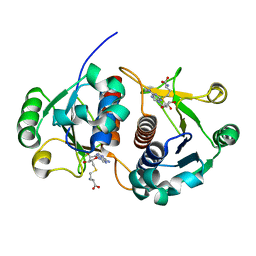 | | Methyltransferase Domain of Human TAR (HIV-1) RNA binding protein 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TAR (HIV-1) RNA loop binding protein | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Battaile, K, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Methyltransferase Domain of Human TAR (HIV-1) RNA binding protein 1 in complex with S-adenosyl-L-homocystein.
To be Published
|
|
7ESR
 
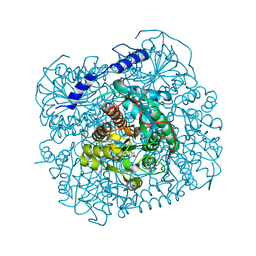 | |
3PVF
 
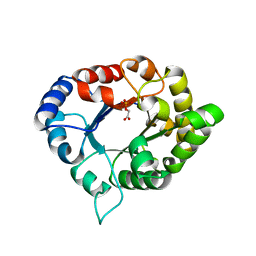 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
6JH7
 
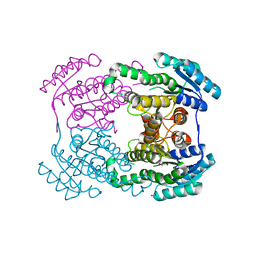 | | Crystal structure of AerF from Microcystis aeruginosa | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Short chain dehydrogenase family protein, ... | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
6JHB
 
 | |
6JHA
 
 | | Crystal structure of NADPH bound AerF from Microcystis aeruginosa | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase family protein, TRIETHYLENE GLYCOL | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
1MI8
 
 | | 2.0 Angstrom crystal structure of a DnaB intein from Synechocystis sp. PCC 6803 | | Descriptor: | DnaB intein | | Authors: | Ding, Y, Chen, X, Ferrandon, S, Xu, M, Rao, Z. | | Deposit date: | 2002-08-22 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing
J.Biol.Chem., 278, 2003
|
|
3PWA
 
 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
4K00
 
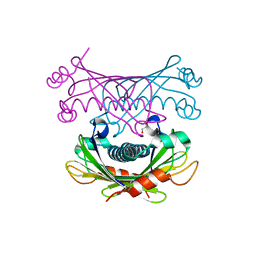 | | Crystal structure of Slr0204, a 1,4-dihydroxy-2-naphthoyl-CoA thioesterase from Synechocystis | | Descriptor: | 1,2-ETHANEDIOL, 1,4-dihydroxy-2-naphthoyl-CoA hydrolase | | Authors: | Furt, F, Allen, W.J, Widhalm, J.R, Madzelan, P, Rizzo, R.C, Basset, G, Wilson, M.A. | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional convergence of structurally distinct thioesterases from cyanobacteria and plants involved in phylloquinone biosynthesis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6K36
 
 | |
3PY2
 
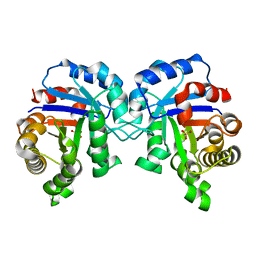 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
1S2O
 
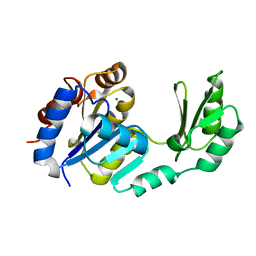 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | | Descriptor: | MAGNESIUM ION, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.L. | | Deposit date: | 2004-01-09 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1TJ3
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in a closed conformation | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1TJ4
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
