5XG8
 
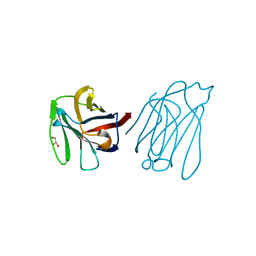 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | GLYCEROL, Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J.Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
5XG7
 
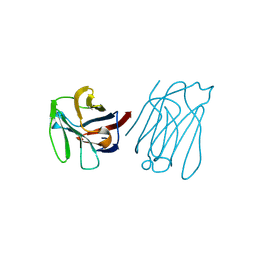 | | Galectin-13/Placental Protein 13 crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y, Wang, Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
5Y03
 
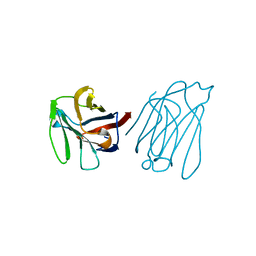 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138.
Sci Rep, 8, 2018
|
|
1P9F
 
 | |
2WY1
 
 | |
2WXW
 
 | |
2WXX
 
 | |
2WY0
 
 | | Crystal structure of mouse angiotensinogen in the oxidised form with space group P6122 | | Descriptor: | 1,2-ETHANEDIOL, ANGIOTENSINOGEN, SODIUM ION | | Authors: | Zhou, A, Wei, Z, Carrell, R.W, Read, R.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A Redox Switch in Angiotensinogen Modulates Angiotensin Release.
Nature, 468, 2010
|
|
2WXZ
 
 | |
2X0B
 
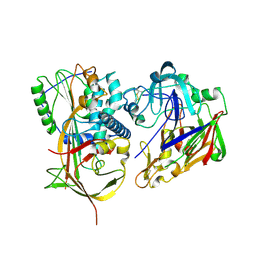 | | Crystal structure of human angiotensinogen complexed with renin | | Descriptor: | ANGIOTENSINOGEN, RENIN | | Authors: | Zhou, A, Wei, Z, Yan, Y, Carrell, R.W, Read, R.J. | | Deposit date: | 2009-12-08 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.33 Å) | | Cite: | A Redox Switch in Angiotensinogen Modulates Angiotensin Release.
Nature, 468, 2010
|
|
2WXY
 
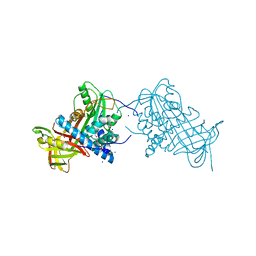 | | Crystal structure of mouse angiotensinogen in the reduced form | | Descriptor: | 1,2-ETHANEDIOL, ANGIOTENSINOGEN, SODIUM ION | | Authors: | Zhou, A, Wei, Z, Carrell, R.W, Read, R.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Redox Switch in Angiotensinogen Modulates Angiotensin Release.
Nature, 468, 2010
|
|
6A62
 
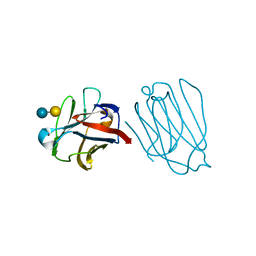 | | Placental protein 13/galectin-13 variant R53HH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A64
 
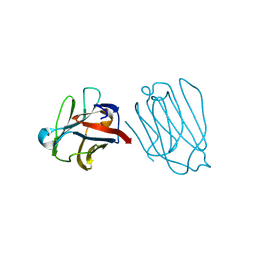 | | Placental protein 13/galectin-13 variant R53HR55NH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A65
 
 | | Placental protein 13/galectin-13 variant R53HR55N with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A63
 
 | | Placental protein 13/galectin-13 variant R53HH57R with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A66
 
 | | Placental protein 13/galectin-13 variant R53H with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
2K7F
 
 | | HADDOCK calculated model of the complex between the BRCT region of RFC p140 and dsDNA | | Descriptor: | 5'-D(P*DCP*DGP*DAP*DCP*DCP*DTP*DCP*DGP*DAP*DGP*DAP*DTP*DCP*DA)-3', 5'-D(P*DCP*DTP*DCP*DGP*DAP*DGP*DGP*DTP*DCP*DG)-3', Replication factor C subunit 1 | | Authors: | Kobayashi, M, Ab, E, Bonvin, A, Siegal, G. | | Deposit date: | 2008-08-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex.
J.Biol.Chem., 285, 2010
|
|
1LA3
 
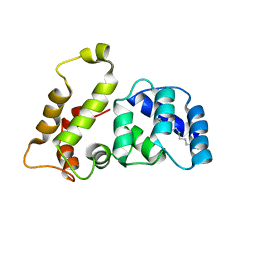 | | Solution structure of recoverin mutant, E85Q | | Descriptor: | CALCIUM ION, MYRISTIC ACID, Recoverin | | Authors: | Ames, J.B, Hamasaki, N, Molchanova, T. | | Deposit date: | 2002-03-27 | | Release date: | 2002-06-19 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state.
Biochemistry, 41, 2002
|
|
1LOB
 
 | |
1LOA
 
 | |
