1BAS
 
 | |
1NIP
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
102L
 
 | |
104L
 
 | |
103L
 
 | |
1CLL
 
 | |
1HRI
 
 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
1GMP
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMR
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMQ
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1PIP
 
 | | CRYSTAL STRUCTURE OF PAPAIN-SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE COMPLEX AT 1.7 ANGSTROMS RESOLUTION: NONCOVALENT BINDING MODE OF A COMMON SEQUENCE OF ENDOGENOUS THIOL PROTEASE INHIBITORS | | Descriptor: | Papain, SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE | | Authors: | Yamamoto, A, Tomoo, K, Doi, M, Ohishi, H, Inoue, M, Ishida, T, Yamamoto, D, Tsuboi, S, Okamoto, H, Okada, Y. | | Deposit date: | 1992-10-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papain-succinyl-Gln-Val-Val-Ala-Ala-p-nitroanilide complex at 1.7-A resolution: noncovalent binding mode of a common sequence of endogenous thiol protease inhibitors.
Biochemistry, 31, 1992
|
|
1OVB
 
 | | THE MECHANISM OF IRON UPTAKE BY TRANSFERRINS: THE STRUCTURE OF AN 18KD NII-DOMAIN FRAGMENT AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, OVOTRANSFERRIN | | Authors: | Kuser, P, Lindley, P, Sarra, R. | | Deposit date: | 1992-10-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of iron uptake by transferrins: the structure of an 18 kDa NII-domain fragment from duck ovotransferrin at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1TRD
 
 | |
1TRE
 
 | |
1DTC
 
 | |
1FBC
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1IZB
 
 | |
1IZA
 
 | |
1FBD
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBH
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBE
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, ZINC ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBF
 
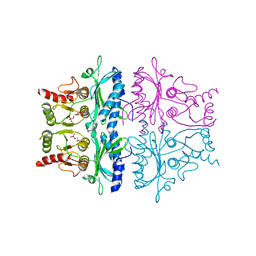 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBG
 
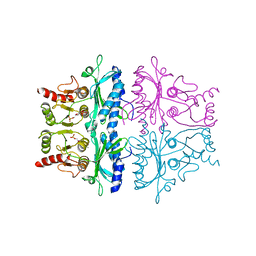 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1DA2
 
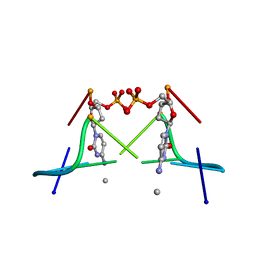 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO4CG): N4-METHOXYCYTOSINE/GUANINE BASE-PAIRS IN Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C45)P*G)-3') | | Authors: | Van Meervelt, L, Moore, M.H, Lin, P.K.T, Brown, D.M, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA.
J.Mol.Biol., 216, 1990
|
|
1D96
 
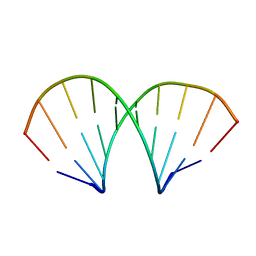 | | MOLECULAR STRUCTURE OF R(GCG)D(TATACGC): A DNA-RNA HYBRID HELIX JOINED TO DOUBLE HELICAL DNA | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP*)-D(*TP*AP*TP*AP*CP*GP*C)-3') | | Authors: | Wang, A.H.-J, Fujii, S, Van Boom, J.H, Van Der Marel, G.A, Van Boeckel, S.A.A, Rich, A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA.
Nature, 299, 1982
|
|
