3UMV
 
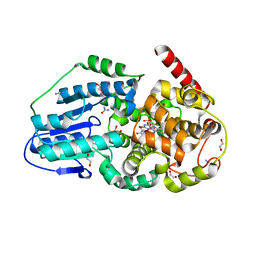 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
3IGI
 
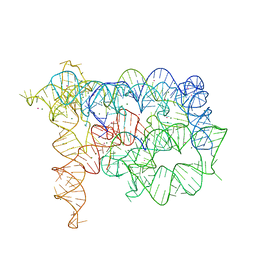 | | Tertiary Architecture of the Oceanobacillus Iheyensis Group II Intron | | Descriptor: | 5'-R(*CP*GP*CP*UP*CP*UP*AP*CP*UP*CP*UP*AP*U)-3', Group IIC intron, MAGNESIUM ION, ... | | Authors: | Toor, N, Keating, K.S, Fedorova, O, Rajashankar, K, Wang, J, Pyle, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | Tertiary architecture of the Oceanobacillus iheyensis group II intron.
Rna, 16, 2010
|
|
6LVB
 
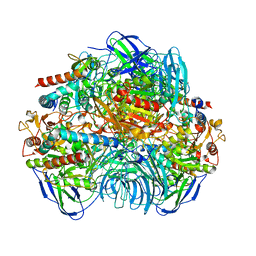 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5X8D
 
 | |
5X8K
 
 | |
5X8B
 
 | | Crystal structure of ATP-TMP and ADP bound thymidylate kinase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5X8V
 
 | |
5X98
 
 | |
5X86
 
 | | Crystal structure of TMP bound thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5XAL
 
 | |
5XT8
 
 | | Magnesium bound apo structure of thymidylate kinase (form I) from Thermus thermophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-06-17 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3NA5
 
 | | Crystal structure of a bacterial phosphoglucomutase, an enzyme important in the virulence of several human pathogens. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Mehra-Chaudhary, R, Beamer, L.J. | | Deposit date: | 2010-06-01 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens.
Proteins, 79, 2011
|
|
3RJG
 
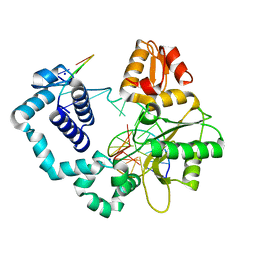 | | Binary complex of DNA Polymerase Beta with a gapped DNA containing 8odG:dA base-pair at primer Terminus | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*(8OG)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5X7J
 
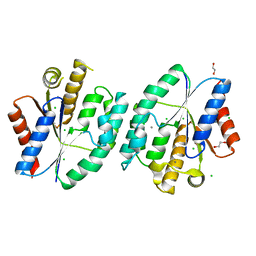 | | Crystal structure of thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2019
|
|
6LVD
 
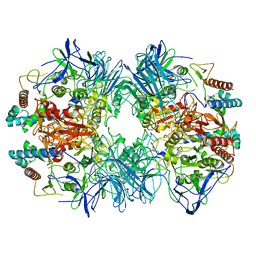 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVV
 
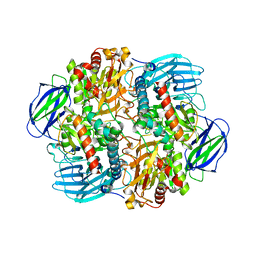 | | N, N-dimethylformamidase | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N-dimethylformamidase large subunit, ... | | Authors: | Arya, C.K, Ramaswamy, S, Kutti, R.V, Gurunath, R. | | Deposit date: | 2020-02-05 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4OY2
 
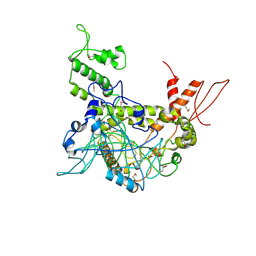 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | Authors: | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1GDD
 
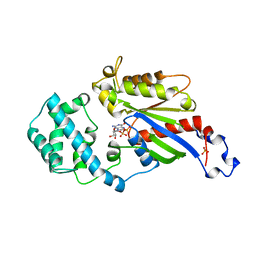 | |
1ANG
 
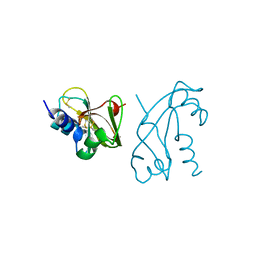 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN REVEALS THE STRUCTURAL BASIS FOR ITS FUNCTIONAL DIVERGENCE FROM RIBONUCLEASE | | Descriptor: | ANGIOGENIN | | Authors: | Acharya, K.R, Allen, S, Shapiro, R, Riordan, J.F, Vallee, B.L. | | Deposit date: | 1994-01-18 | | Release date: | 1995-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
5F24
 
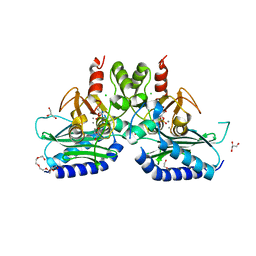 | | Crystal structure of dual specific IMPase/NADP phosphatase bound with D-inositol-1-phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYH
 
 | | Crystal Structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ at pH 7.0 | | Descriptor: | CALCIUM ION, GLYCEROL, Inositol monophosphatase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYG
 
 | | Crystal structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3QMF
 
 | | Crystal strucuture of an inositol monophosphatase family protein (SAS2203) from Staphylococcus aureus MSSA476 | | Descriptor: | Inositol monophosphatase family protein, SULFATE ION | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2011-02-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
3RYD
 
 | | Crystal structure of Ca bound IMPase family protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Inositol monophosphatase family protein, POTASSIUM ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
1F17
 
 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
