1IHK
 
 | | CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 9 (FGF9) | | Descriptor: | GLIA-ACTIVATING FACTOR, PHOSPHATE ION | | Authors: | Plotnikov, A.N, Eliseenkova, A.V, Ibrahimi, O.A, Lemmon, M.A, Mohammadi, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition.
J.Biol.Chem., 276, 2001
|
|
1II4
 
 | | CRYSTAL STRUCTURE OF SER252TRP APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, HEPARIN-BINDING GROWTH FACTOR 2 | | Authors: | Ibrahimi, O.A, Eliseenkova, A.V, Plotnikov, A.N, Ornitz, D.M, Mohammadi, M. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1SEU
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Indolocarbazole SA315F and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 2,10-DIHYDROXY-12-(BETA-D-GLUCOPYRANOSYL)-6,7,12,13-TETRAHYDROINDOLO[2,3-A]PYRROLO[3,4-C]CARBAZOLE-5,7-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-02-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1ILT
 
 | |
1S6H
 
 | | PORCINE TRYPSIN COMPLEXED WITH GUANIDINE-3-PROPANOL INHIBITOR | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-23 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S4V
 
 | | The 2.0 A crystal structure of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm | | Descriptor: | DVA-LEU-LYS-0QE peptide, SULFATE ION, cysteine endopeptidase | | Authors: | Than, M.E, Helm, M, Simpson, D.J, Lottspeich, F, Huber, R, Gietl, C. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm.
J.Mol.Biol., 336, 2004
|
|
1IN8
 
 | | THERMOTOGA MARITIMA RUVB T158V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IDF
 
 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IOA
 
 | | ARCELIN-5, A LECTIN-LIKE DEFENSE PROTEIN FROM PHASEOLUS VULGARIS | | Descriptor: | ARCELIN-5A, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hamelryck, T, Loris, R. | | Deposit date: | 1996-10-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of arcelin-5, a lectin-like defense protein from Phaseolus vulgaris
J.Biol.Chem., 271, 1996
|
|
1SBW
 
 | | CRYSTAL STRUCTURE OF MUNG BEAN INHIBITOR LYSINE ACTIVE FRAGMENT COMPLEX WITH BOVINE BETA-TRYPSIN AT 1.8A RESOLUTION | | Descriptor: | CALCIUM ION, PROTEIN (BETA-TRYPSIN), PROTEIN (MUNG BEAN INHIBITOR LYSIN ACTIVE FRAGMENT), ... | | Authors: | Huang, Q, Zhu, Y, Chi, C, Tang, Y. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mung bean inhibitor lysine active fragment complex with bovine beta-trypsin at 1.8A resolution.
J.Biomol.Struct.Dyn., 16, 1999
|
|
1IQQ
 
 | | Crystal Structure of Japanese pear S3-RNase | | Descriptor: | S3-RNase, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Matsuura, T, Sakai, H, Norioka, S. | | Deposit date: | 2001-07-25 | | Release date: | 2001-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility.
J.Biol.Chem., 276, 2001
|
|
1SBX
 
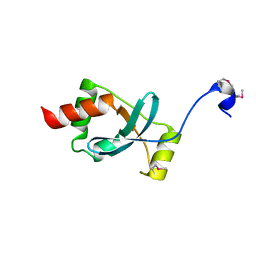 | | Crystal structure of the Dachshund-homology domain of human SKI | | Descriptor: | Ski oncogene | | Authors: | Wilson, J.J, Malakhova, M, Zhang, R, Joachimiak, A, Hegde, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Dachshund Homology Domain of human SKI
Structure, 12, 2004
|
|
1IR3
 
 | |
1SE4
 
 | | STAPHYLOCOCCAL ENTEROTOXIN B COMPLEXED WITH LACTOSE | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN B, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Swaminathan, S, Sax, M. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Residues defining V beta specificity in staphylococcal enterotoxins.
Nat.Struct.Biol., 2, 1995
|
|
1SCE
 
 | |
1SCQ
 
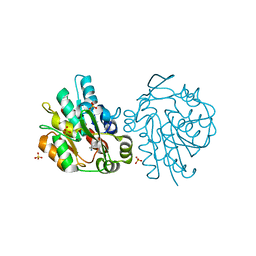 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1J6O
 
 | |
1SEF
 
 | |
1II2
 
 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase (PEPCK) from Trypanosoma cruzi | | Descriptor: | PHOSPHOENOLPYRUVATE CARBOXYKINASE, SULFATE ION | | Authors: | Trapani, S, Linss, J, Goldenberg, S, Fischer, H, Craievich, A.F, Oliva, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the dimeric phosphoenolpyruvate carboxykinase (PEPCK) from Trypanosoma cruzi at 2 A resolution.
J.Mol.Biol., 313, 2001
|
|
1J84
 
 | | STRUCTURE OF FAM17 CARBOHYDRATE BINDING MODULE FROM CLOSTRIDIUM CELLULOVORANS WITH BOUND CELLOTETRAOSE | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta glucanase EngF | | Authors: | Notenboom, V, Boraston, A.B, Chiu, P, Freelove, A.C.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-05-20 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study.
J.Mol.Biol., 314, 2001
|
|
1RXO
 
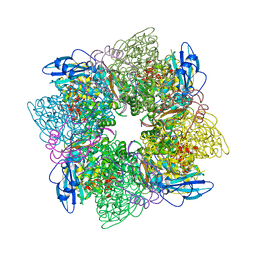 | | ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATE AND CALCIUM | | Descriptor: | CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-12-06 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate.
J.Mol.Biol., 265, 1997
|
|
1IJ2
 
 | |
1SHG
 
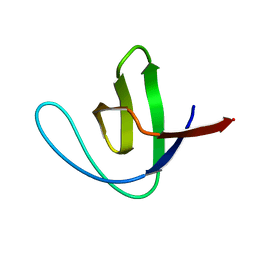 | | CRYSTAL STRUCTURE OF A SRC-HOMOLOGY 3 (SH3) DOMAIN | | Descriptor: | ALPHA-SPECTRIN SH3 DOMAIN | | Authors: | Noble, M, Pauptit, R, Musacchio, A, Saraste, M, Wierenga, R.K. | | Deposit date: | 1993-05-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Src-homology 3 (SH3) domain.
Nature, 359, 1992
|
|
1SHR
 
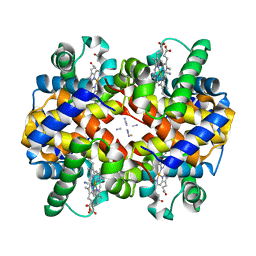 | | Crystal structure of ferrocyanide bound human hemoglobin A2 at 1.88A resolution | | Descriptor: | CYANIDE ION, FE (III) ION, Hemoglobin alpha chain, ... | | Authors: | Sen, U, Dasgupta, J, Choudhury, D, Datta, P, Chakrabarti, A, Chakrabarty, S.B, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2004-02-26 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of HbA2 and HbE and modeling of hemoglobin delta4: interpretation of the thermal stability and the antisickling effect of HbA2 and identification of the ferrocyanide binding site in Hb
Biochemistry, 43, 2004
|
|
1IEB
 
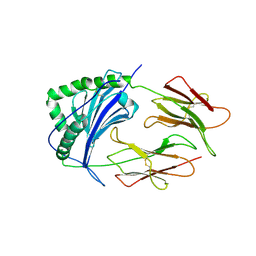 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-EK, SULFATE ION | | Authors: | Fremont, D.H, Hendrickson, W.A, Marrack, P, Kappler, J. | | Deposit date: | 1996-04-05 | | Release date: | 1997-06-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of an MHC class II molecule with covalently bound single peptides.
Science, 272, 1996
|
|
