8HI6
 
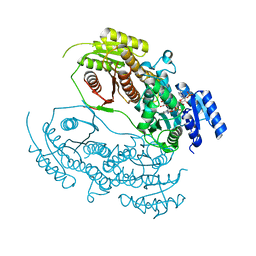 | |
8H9E
 
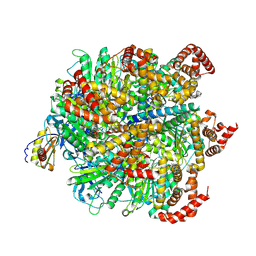 | | Human ATP synthase F1 domain, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
1RDD
 
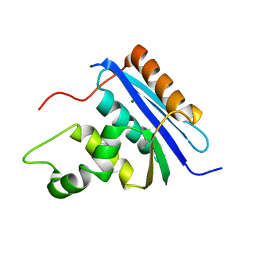 | |
5L5E
 
 | |
8HM0
 
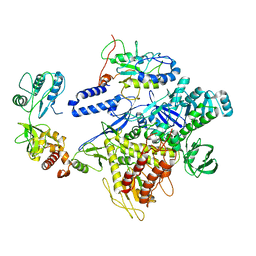 | | F8-A22-E4 complex of MPXV in trimeric form | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Li, Y.N, Shen, Y.P, Hu, Z.W, Yan, R.H. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the assembly of the DNA polymerase holoenzyme from a monkeypox virus variant.
Sci Adv, 9, 2023
|
|
8H9M
 
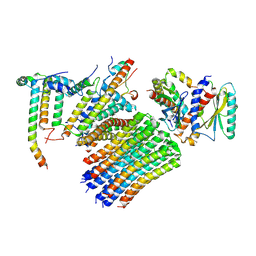 | | Human ATP synthase state 3a subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
5L5Y
 
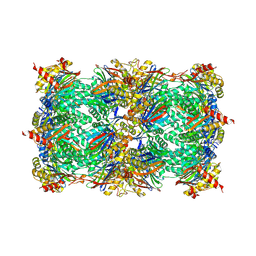 | |
4M9S
 
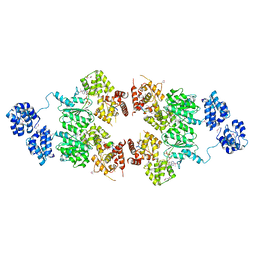 | | crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
1RE0
 
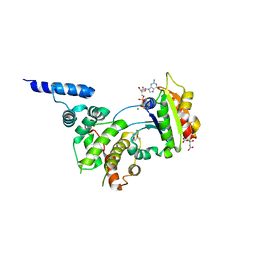 | | Structure of ARF1-GDP bound to Sec7 domain complexed with Brefeldin A | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-ribosylation factor 1, ARF guanine-nucleotide exchange factor 1, ... | | Authors: | Goldberg, J, Mossessova, E. | | Deposit date: | 2003-11-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism.
Mol.Cell, 12, 2003
|
|
1R5V
 
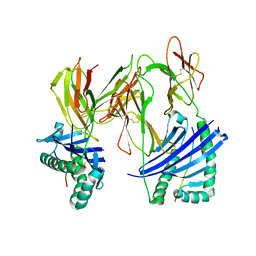 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E.J, He, X.L, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation.
Mol.Cell, 12, 2003
|
|
8H9S
 
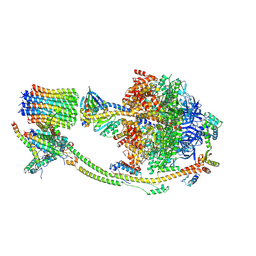 | | Human ATP synthase state 1 (combined) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
1RE2
 
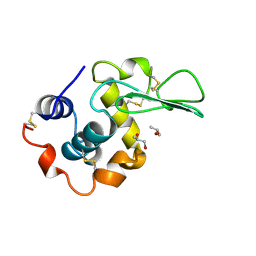 | | HUMAN LYSOZYME LABELLED WITH TWO 2',3'-EPOXYPROPYL BETA-GLYCOSIDE OF N-ACETYLLACTOSAMINE | | Descriptor: | GLYCEROL, PROTEIN (LYSOZYME), beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1998-11-05 | | Release date: | 1999-05-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual affinity labeling of the active site of human lysozyme with an N-acetyllactosamine derivative: first ligand assisted recognition of the second ligand.
Biochemistry, 38, 1999
|
|
2B8S
 
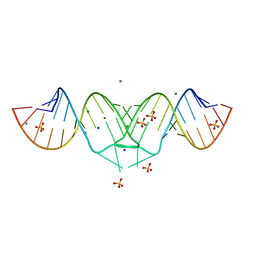 | | Structure of HIV-1(MAL) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
NAT.STRUCT.BIOL., 8, 2001
|
|
8H9K
 
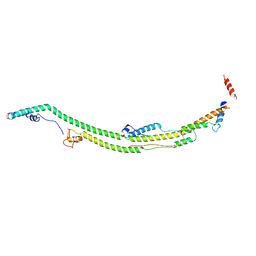 | | Human ATP synthase state 2 subregion 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase subunit d, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
2MUK
 
 | |
1R6B
 
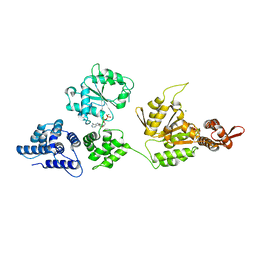 | | High resolution crystal structure of ClpA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ClpA protein, MAGNESIUM ION | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
8H9G
 
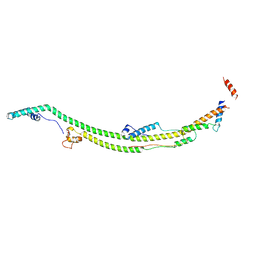 | | Human ATP synthase state 1 subregion 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase subunit d, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9L
 
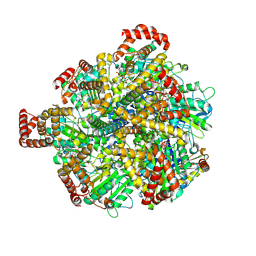 | | Human ATP synthase F1 domain, state 3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
1T1N
 
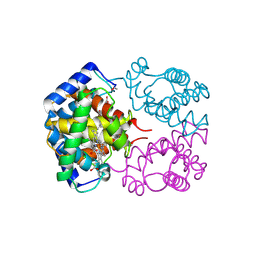 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Vitagliano, L, Savino, C, Zagari, A. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trematomus newnesi haemoglobin re-opens the root effect question.
J.Mol.Biol., 287, 1999
|
|
8H9J
 
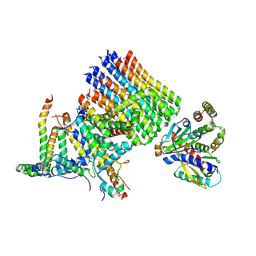 | | Human ATP synthase state2 subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9T
 
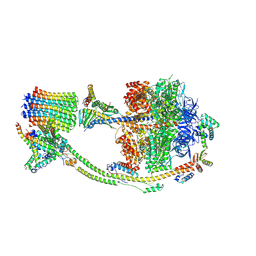 | | Human ATP synthase state 2 (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
2N1K
 
 | |
1R76
 
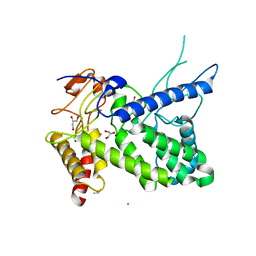 | | Structure of a pectate lyase from Azospirillum irakense | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Novoa de Armas, H, Verboven, C, De Ranter, C, Desair, J, Vande Broek, A, Vanderleyden, J, Rabijns, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Azospirillum irakense pectate lyase displays a toroidal fold.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8HLZ
 
 | | F8-A22-E4 complex of MPXV in hexameric form | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Li, Y.N, Shen, Y.P, Hu, Z.W, Yan, R.H. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the assembly of the DNA polymerase holoenzyme from a monkeypox virus variant.
Sci Adv, 9, 2023
|
|
2AIH
 
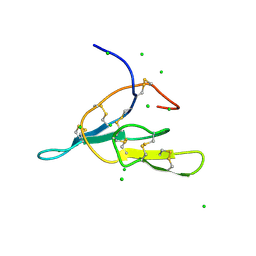 | | 1H-NMR solution structure of a trypsin/chymotrypsin Bowman-Birk inhibitor from Lens culinaris. | | Descriptor: | Bowman-Birk type protease inhibitor, LCTI, CHLORIDE ION | | Authors: | Ragg, E.M, Galbusera, V, Scarafoni, A, Negri, A, Tedeschi, G, Consonni, A, Sessa, F, Duranti, M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-08-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Inhibitory properties and solution structure of a potent Bowman-Birk protease inhibitor from lentil (Lens culinaris, L) seeds.
Febs J., 273, 2006
|
|
