2HUU
 
 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase in complex with alanine | | Descriptor: | 1-BUTANOL, ALANINE, Alanine glyoxylate aminotransferase | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
2HYP
 
 | | Crystal structure of Rv0805 D66A mutant | | Descriptor: | BROMIDE ION, CACODYLATE ION, FE (III) ION, ... | | Authors: | Shenoy, A.R, Capuder, M, Draskovic, P, Lamba, D, Visweswariah, S.S, Podobnik, M. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Biochemical Analysis of the Rv0805 Cyclic Nucleotide Phosphodiesterase from Mycobacterium tuberculosis.
J.Mol.Biol., 365, 2007
|
|
2HYX
 
 | |
2G0I
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
2FY5
 
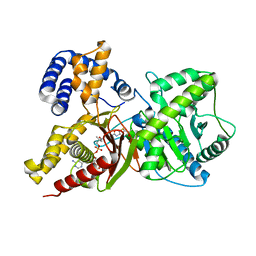 | | Structures of ligand bound human choline acetyltransferase provide insight into regulation of acetylcholine synthesis | | Descriptor: | Choline O-acetyltransferase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-2,2-DIMETHYL-4-OXO-4-{[3-OXO-3-({2-[(2-OXOPROPYL)THIO]ETHYL}AMINO)PROPYL]AMINO}BUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Kim, A.R, Rylett, R.J, Shilton, B.H. | | Deposit date: | 2006-02-07 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate binding and catalytic mechanism of human choline acetyltransferase.
Biochemistry, 45, 2006
|
|
2FYM
 
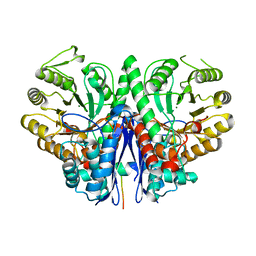 | |
2G2U
 
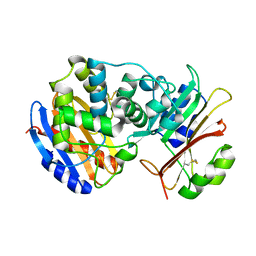 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2GF4
 
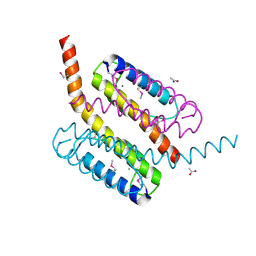 | | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14 | | Descriptor: | ACETATE ION, CALCIUM ION, Protein Vng1086c | | Authors: | Benach, J, Zhou, W, Jayaraman, S, Forouhar, F.F, Janjua, H, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14
To be Published
|
|
2GA0
 
 | | Variable Small Protein 1 of Borrelia turicatae (VspA or Vsp1) | | Descriptor: | NICKEL (II) ION, surface protein VspA | | Authors: | Lawson, C.L, Yung, B.H, Barbour, A.G, Zuckert, W.R. | | Deposit date: | 2006-03-07 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of neurotropism-associated variable surface protein 1 (Vsp1) of Borrelia turicatae.
J.Bacteriol., 188, 2006
|
|
2GDE
 
 | | Thrombin in complex with inhibitor | | Descriptor: | (R)-3-((2S,3R)-1-((2S,3AR,5S,6S,7AS)-2-(2-(1-CARBAMIMIDOYL-2,5-DIHYDRO-1H-PYRROL-3-YL)ETHYLCARBAMOYL)-5,6-DIHYDROXYOCTAHYDRO-1H-INDOL-1-YL)-3-CHLORO-4-METHYL-1-OXOPENTAN-2-YLAMINO)-2-METHOXY-3-OXOPROPYL HYDROGEN SULFATE, Hirudin, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2006-03-16 | | Release date: | 2007-03-20 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Total synthesis and structural confirmation of chlorodysinosin A.
J.Am.Chem.Soc., 128, 2006
|
|
2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
2FYQ
 
 | |
2G60
 
 | | Structure of anti-FLAG M2 Fab domain | | Descriptor: | anti-FLAG M2 Fab heavy chain, anti-FLAG M2 Fab light chain | | Authors: | Roosild, T.P. | | Deposit date: | 2006-02-23 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of anti-FLAG M2 Fab domain and its use in the stabilization of engineered membrane proteins.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GC0
 
 | |
2G7Y
 
 | | Human Cathepsin S with inhibitor CRA-16981 | | Descriptor: | (1R)-2-[(CYANOMETHYL)AMINO]-1-({[2-(DIFLUOROMETHOXY)BENZYL]SULFONYL}METHYL)-2-OXOETHYL MORPHOLINE-4-CARBOXYLATE, Cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-03-01 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human Cathepsin S with inhibitor CRA-16981
To be Published
|
|
2FY3
 
 | |
1EEF
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER COMPLEXED WITH BOUND LIGAND PEPG | | Descriptor: | 2-PHENETHYL-2,3-DIHYDRO-PHTHALAZINE-1,4-DIONE, PROTEIN (HEAT-LABILE ENTEROTOXIN B CHAIN), alpha-D-galactopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ERT
 
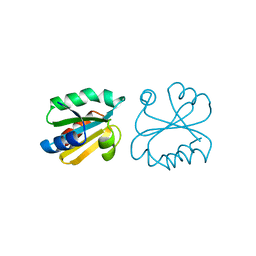 | | HUMAN THIOREDOXIN (REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Weichsel, A, Gasdaska, J.R, Powis, G, Montfort, W.R. | | Deposit date: | 1996-02-07 | | Release date: | 1996-10-14 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer.
Structure, 4, 1996
|
|
3D93
 
 | |
1ESC
 
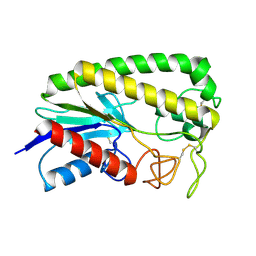 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
3DOB
 
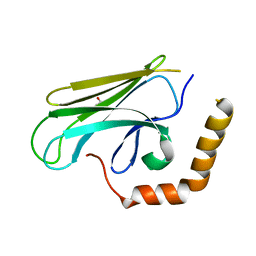 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
1EFD
 
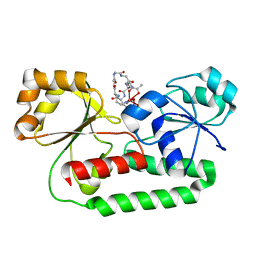 | | PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH GALLICHROME | | Descriptor: | FERRICHROME-BINDING PERIPLASMIC PROTEIN, GALLICHROME | | Authors: | Clarke, T.E, Ku, S.-Y, Dougan, D.R, Vogel, H.J, Tari, L.W. | | Deposit date: | 2000-02-07 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the ferric siderophore binding protein FhuD complexed with gallichrome.
Nat.Struct.Biol., 7, 2000
|
|
1ESQ
 
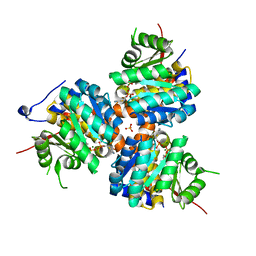 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) WITH ATP AND THIAZOLE PHOSPHATE. | | Descriptor: | 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, HYDROXYETHYLTHIAZOLE KINASE, ... | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1EFS
 
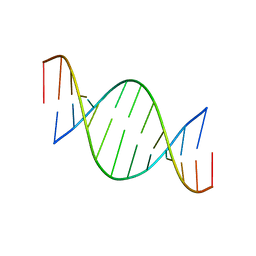 | | CONFORMATION OF A DNA-RNA HYBRID | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*UP*CP*UP*UP*CP*CP*UP*CP*UP*C)-3') | | Authors: | Larue, V, Hantz, E. | | Deposit date: | 2000-02-10 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of an RNA--DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand.
Int.J.Biol.Macromol., 28, 2001
|
|
3D9V
 
 | |
