2ZOY
 
 | | The multi-drug binding transcriptional repressor CgmR (CGL2612 protein) from C.glutamicum | | Descriptor: | GLYCEROL, Transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CGL2612 protein from Corynebacterium glutamicum is a drug resistance-related transcriptional repressor: structural and functional analysis of a newly identified transcription factor from genomic DNA analysis
J.Biol.Chem., 280, 2005
|
|
3EFF
 
 | | The Crystal Structure of Full-Length KcsA in its Closed Conformation | | Descriptor: | FAB, Voltage-gated potassium channel | | Authors: | Uysal, S, Vasquez, V, Tereshko, T, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6NVO
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
2ZB7
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADPH and nicotinamide | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
4PL1
 
 | |
1JXO
 
 | |
5AVU
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 7.0 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW2
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 85 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVT
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW3
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 100 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
2DBU
 
 | |
1TOC
 
 | |
4P9F
 
 | | E. coli McbR/YncC | | Descriptor: | HTH-type transcriptional regulator mcbR | | Authors: | Lord, D.M, Page, R, Peti, W. | | Deposit date: | 2014-04-03 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | McbR/YncC: Implications for the Mechanism of Ligand and DNA Binding by a Bacterial GntR Transcriptional Regulator Involved in Biofilm Formation.
Biochemistry, 53, 2014
|
|
2DBW
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Acyl-Enzyme Intermediate | | Descriptor: | GAMMA-L-GLUTAMIC ACID, GLYCEROL, Gamma-glutamyltranspeptidase | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2DDT
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with magnesium ion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ago, H, Oda, M, Tsuge, H, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DBX
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Complexed with L-Glutamate | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5AVR
 
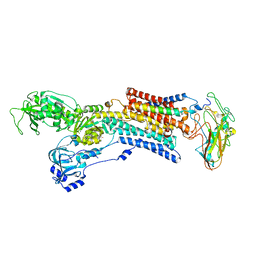 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 1.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVZ
 
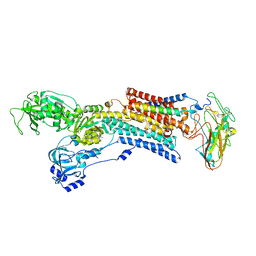 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 55 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVW
 
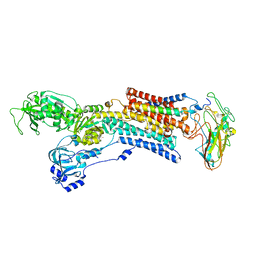 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 16.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW8
 
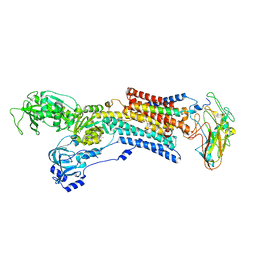 | | Kinetics by X-ray crystallography: E2.MgF42-.2RB+ crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
2ZXO
 
 | | Crystal structure of RecJ from Thermus thermophilus HB8 | | Descriptor: | Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
1K7G
 
 | | PrtC from Erwinia chrysanthemi | | Descriptor: | CALCIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Hege, T, Baumann, U. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1K7I
 
 | | PrtC from Erwinia chrysanthemi: Y228F mutant | | Descriptor: | CALCIUM ION, ZINC ION, secreted protease C | | Authors: | Baumann, U, Hege, T. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1U5M
 
 | | Structure of a Chordin-like Cysteine-rich Repeat (VWC module) from Collagen IIA | | Descriptor: | alpha 1 type II collagen isoform 1 | | Authors: | O'Leary, J.M, Hamilton, J.m, Deane, C.M, Valeyev, N.V, Sandell, L.J, Downing, A.K. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-05 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA
J.Biol.Chem., 279, 2004
|
|
1KAM
 
 | | Structure of Bacillus subtilis Nicotinic Acid Mononucleotide Adenylyl Transferase | | Descriptor: | NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE | | Authors: | Olland, A.M, Underwood, K.W, Czerwinski, R.M, Lo, M.C, Aulabaugh, A, Bard, J, Stahl, M.L, Somers, W.S, Sullivan, F.X, Chopra, R. | | Deposit date: | 2001-11-02 | | Release date: | 2002-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase.
J.Biol.Chem., 277, 2002
|
|
