1QPH
 
 | |
5GKC
 
 | | The crystal structure of the CPS-6 H148A/F122A | | Descriptor: | Endonuclease G, mitochondrial | | Authors: | Lin, J.L, Yuan, H.S. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of endonuclease G in complex with DNA reveals how it nonspecifically degrades DNA as a homodimer.
Nucleic Acids Res., 44, 2016
|
|
6EVU
 
 | | Adhesin domain of PrgB from Enterococcus faecalis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, PrgB | | Authors: | Schmitt, A, Berntsson, R.P.A. | | Deposit date: | 2017-11-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction.
Mol. Microbiol., 109, 2018
|
|
2B0D
 
 | | EcoRV Restriction Endonuclease/GAATTC/Ca2+ | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*AP*TP*TP*CP*TP*T)-3', CALCIUM ION, Type II restriction enzyme EcoRV | | Authors: | Hiller, D.A, Rodriguez, A.M, Perona, J.J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-cognate Enzyme-DNA Complex: Structural and Kinetic Analysis of EcoRV Endonuclease Bound to the EcoRI Recognition Site GAATTC
J.Mol.Biol., 354, 2005
|
|
5CR5
 
 | |
6RY3
 
 | | Structure of the WUS homeodomain | | Descriptor: | ACETYL GROUP, Protein WUSCHEL, SULFATE ION | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
7WJ0
 
 | |
7NDY
 
 | | Di-phosphorylated Barrier-to-Autointegration Factor (BAF) in complex with LEM domain of Emerin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Barrier-to-autointegration factor, N-terminally processed, ... | | Authors: | Marcelot, A, Le Du, M.H, Hoffmann, G, Zinn-Justin, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
4RXJ
 
 | | crystal structure of WHSC1L1-PWWP2 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
6DPN
 
 | |
1RZ9
 
 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | Descriptor: | 26-MER, Rep protein | | Authors: | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
3VDP
 
 | |
2Z6K
 
 | | Crystal structure of full-length human RPA14/32 heterodimer | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Habel, J.E, Kabaleeswaran, V, Borgstahl, G.E. | | Deposit date: | 2007-08-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation
J.Mol.Biol., 374, 2007
|
|
3VE5
 
 | |
3JXB
 
 | |
3JXD
 
 | |
4YRV
 
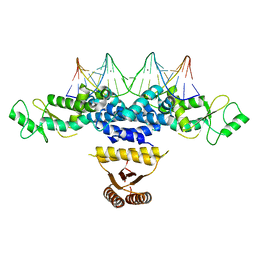 | | Crystal structure of Anabaena transcription factor HetR complexed with 21-bp DNA from hetP promoter | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Zhang, C.C, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-03-16 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into HetR-PatS interaction involved in cyanobacterial pattern formation
Sci Rep, 5, 2015
|
|
6JYK
 
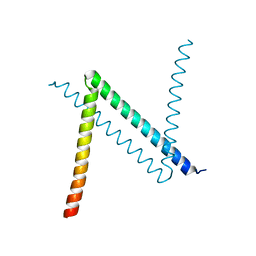 | | Crystal Structure of C. crescentus free GapR | | Descriptor: | UPF0335 protein CCNA_03428 | | Authors: | Xia, B, Huang, Q. | | Deposit date: | 2019-04-26 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GapR binds DNA through dynamic opening of its tetrameric interface.
Nucleic Acids Res., 48, 2020
|
|
1ZEZ
 
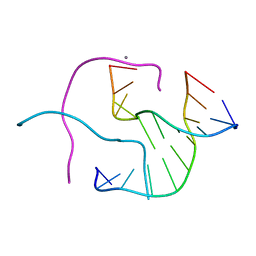 | | ACC Holliday Junction | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6DPI
 
 | |
4ENM
 
 | |
3JXC
 
 | |
5O6F
 
 | | NMR structure of cold shock protein A from Corynebacterium pseudotuberculosis | | Descriptor: | Cold-shock protein | | Authors: | Caruso, I.P, Panwalkar, V, Coronado, M.A, Dingley, A.J, Cornelio, M.L, Willbold, D, Arni, R.K, Eberle, R.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction of Corynebacterium pseudotuberculosis cold shock protein A with Y-box single-stranded DNA fragment.
FEBS J., 285, 2018
|
|
4ENK
 
 | |
3VDU
 
 | |
