5P1X
 
 | |
2R3K
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-5-phenyl-N-(pyrimidin-5-ylmethyl)pyrazolo[1,5-a]pyridin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
3FMM
 
 | | P38 kinase crystal structure in complex with RO6226 | | Descriptor: | 6-(2,4-difluorophenoxy)-N-[(1R)-1-methyl-2-(methylsulfonyl)ethyl]-1H-pyrazolo[3,4-d]pyrimidin-3-amine, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Ghate, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P38 kinase crystal structure in complex with RO6226
To be Published
|
|
5P2B
 
 | |
5P2N
 
 | |
5P32
 
 | |
2CS0
 
 | |
5P3F
 
 | |
1SI5
 
 | | Protease-like domain from 2-chain hepatocyte growth factor | | Descriptor: | hepatocyte growth factor | | Authors: | Kirchhofer, D, Yao, X, Peek, M, Eigenbrot, C, Lipari, M.T, Billeci, K.L, Maun, H.R, Moran, P, Santell, L, Lazarus, R.A. | | Deposit date: | 2004-02-27 | | Release date: | 2004-12-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling
J.Biol.Chem., 279, 2004
|
|
5OZM
 
 | |
5P3V
 
 | |
5P00
 
 | |
1I07
 
 | | EPS8 SH3 DOMAIN INTERTWINED DIMER | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR KINASE SUBSTRATE EPS8 | | Authors: | Kishan, K.V.R, Newcomer, M.E. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain.
Protein Sci., 10, 2001
|
|
1I04
 
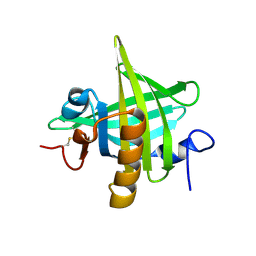 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN-I FROM MOUSE LIVER | | Descriptor: | MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pheromone binding to mouse major urinary protein (MUP-I)
Protein Sci., 10, 2001
|
|
5P4A
 
 | |
1I06
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH SEC-BUTYL-THIAZOLINE | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1X4C
 
 | | Solution structure of RRM domain in splicing factor 2 | | Descriptor: | Splicing factor, arginine/serine-rich 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in splicing factor 2
To be Published
|
|
5P0H
 
 | |
5P4P
 
 | |
1I05
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH HYDROXY-METHYL-HEPTANONE | | Descriptor: | 6-HYDROXY-6-METHYL-HEPTAN-3-ONE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
5P0T
 
 | |
5P54
 
 | |
1X5K
 
 | | The solution structure of the sixth fibronectin type III domain of human Neogenin | | Descriptor: | Neogenin | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the sixth fibronectin type III domain of human Neogenin
To be Published
|
|
5P17
 
 | |
1I0C
 
 | | EPS8 SH3 CLOSED MONOMER | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR KINASE SUBSTRATE EPS8 | | Authors: | Kishan, K.V.R, Newcomer, M.E. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain.
Protein Sci., 10, 2001
|
|
