8USI
 
 | |
8USK
 
 | |
8USL
 
 | |
8USF
 
 | |
4PUD
 
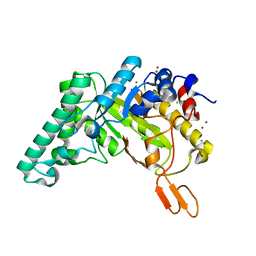 | | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylopentaose in active site | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylopentaose in active site.
To be Published
|
|
3CUG
 
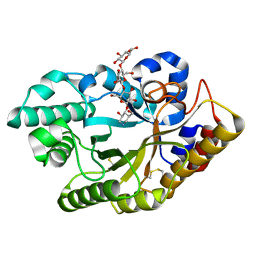 | |
3CUH
 
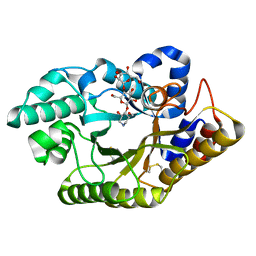 | |
4PMD
 
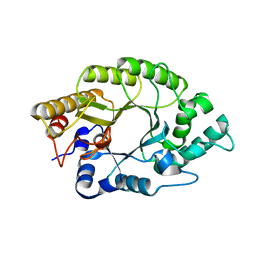 | |
4PRW
 
 | | Xylanase T6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylanase T6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose
To be Published
|
|
3CUF
 
 | |
4QPW
 
 | | BiXyn10A CBM1 with Xylohexaose Bound | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PUE
 
 | | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylotetraose in active site | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylotetraose in active site.
To be Published
|
|
1FH9
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED LACTAM OXIME INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-(2Z,3S,4S,5R)-2-hydroxyiminopiperidine-3,4,5-triol | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1I1W
 
 | | 0.89A Ultra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus | | Descriptor: | ACETONE, ENDO-1,4-BETA-XYLANASE, ETHANOL, ... | | Authors: | Natesh, R, Ramakumar, S, Viswamitra, M.A. | | Deposit date: | 2001-02-04 | | Release date: | 2003-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Thermostable xylanase from Thermoascus aurantiacus at ultrahigh resolution (0.89 A) at 100 K and atomic resolution (1.11 A) at 293 K refined anisotropically to small-molecule accuracy.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FH8
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED ISOFAGOMINE INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1J01
 
 | | Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam | | Descriptor: | (3S,4R)-3-hydroxy-2-oxopiperidin-4-yl beta-D-xylopyranoside, beta-1,4-xylanase | | Authors: | Williams, S.J, Notenboom, V, Wicki, J, Rose, D.R, Withers, S.G. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A New, Simple, High-Affinity Glycosidase Inhibitor: Analysis of Binding through X-ray Crystallography, Mutagenesis, and Kinetic Analysis
J.Am.Chem.Soc., 122, 2000
|
|
1FH7
 
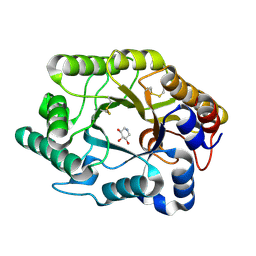 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED INHIBITOR DEOXYNOJIRIMYCIN | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FHD
 
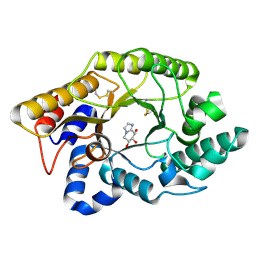 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED IMIDAZOLE INHIBITOR | | Descriptor: | 5,6,7,8-TETRAHYDRO-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, BETA-1,4-XYLANASE, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
2BNJ
 
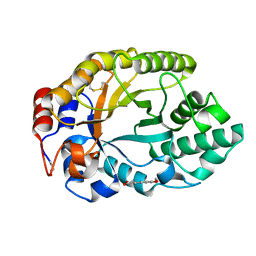 | | The xylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ENDO-1,4-BETA-XYLANASE, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Vardakou, M, Murray, J.W, Flint, J, Christakopoulos, P, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2005-03-25 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family 10 Thermoascus Aurantiacus Xylanase Utilizes Arabinose Decorations of Xylan as Significant Substrate Specificity Determinants.
J.Mol.Biol., 352, 2005
|
|
3W28
 
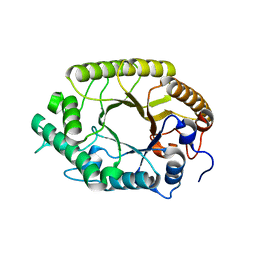 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W29
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W27
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3MSG
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
3W25
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
2DEP
 
 | |
