3GP5
 
 | |
5UM0
 
 | |
5VR6
 
 | | Structure of Human Sts-1 histidine phosphatase domain with sulfate bound | | Descriptor: | SULFATE ION, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
5W5G
 
 | | Structure of Human Sts-1 histidine phosphatase domain | | Descriptor: | Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
3D6A
 
 | | Crystal structure of the 2H-phosphatase domain of Sts-2 in complex with tungstate. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Sts-2 protein, ... | | Authors: | Chen, Y, Carpino, N, Nassar, N. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of the 2H-phosphatase domain of Sts-2 reveals an acid-dependent phosphatase activity.
Biochemistry, 48, 2009
|
|
1C80
 
 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
3D8H
 
 | | Crystal structure of phosphoglycerate mutase from Cryptosporidium parvum, cgd7_4270 | | Descriptor: | Glycolytic phosphoglycerate mutase | | Authors: | Wernimont, A.K, Lew, J, Wasney, G, Alam, Z, Kozieradzki, I, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-23 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of a new phosphatase from Plasmodium.
Mol.Biochem.Parasitol., 179, 2011
|
|
1C7Z
 
 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GLYCERALDEHYDE-3-PHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
3DCY
 
 | |
3DB1
 
 | |
1C81
 
 | | MICHAELIS COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1-deoxy-1-phosphono-6-O-phosphono-D-glucitol, FRUCTOSE-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
2H0Q
 
 | |
1EBB
 
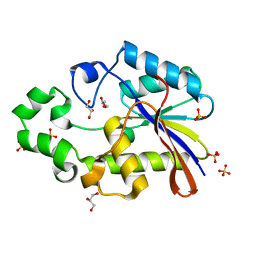 | | Bacillus stearothermophilus YhfR | | Descriptor: | GLYCEROL, PHOSPHATASE, SULFATE ION | | Authors: | Rigden, D.J, Jedrzejas, M.J. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Action of a Cofactor-Dependent Phosphoglycerate Mutase Homolog from Bacillus Stearothermophilus with Broad Specificity Phosphatase Activity.
J.Mol.Biol., 315, 2002
|
|
5VVE
 
 | |
2IKQ
 
 | |
3E9C
 
 | |
5WDI
 
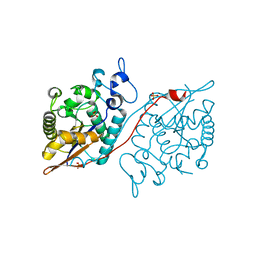 | | Structure of Human Sts-2 histidine phosphatase domain | | Descriptor: | SULFATE ION, Ubiquitin-associated and SH3 domain-containing protein A | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
1E58
 
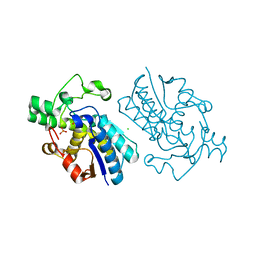 | | E.coli cofactor-dependent phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCERATE MUTASE, SULFATE ION | | Authors: | Bond, C.S, Hunter, W.N. | | Deposit date: | 2000-07-19 | | Release date: | 2001-03-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structure of the Phosphohistidine-Activated Form of Escherichia Coli Cofactor-Dependent Phosphoglycerate Mutase.
J.Biol.Chem., 276, 2001
|
|
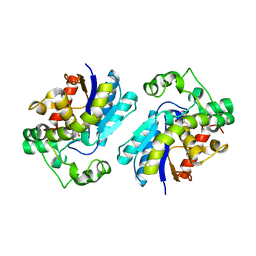
2H4Z
 
 | |
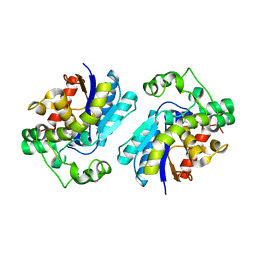
2H4X
 
 | |
3GP3
 
 | |
3FDZ
 
 | | Crystal structure of phosphoglyceromutase from burkholderia pseudomallei 1710b with bound 2,3-diphosphoglyceric acid and 3-phosphoglyceric acid | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3E9D
 
 | | Structure of full-length TIGAR from Danio rerio | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Zgc:56074 | | Authors: | Li, H, Jogl, G. | | Deposit date: | 2008-08-21 | | Release date: | 2008-12-16 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TIGAR (TP53 induced glycolysis and apoptosis regulator) is a fructose-2,6- and fructose-1,6-bisphosphatase
To be Published
|
|
3EOZ
 
 | | Crystal Structure of Phosphoglycerate Mutase from Plasmodium Falciparum, PFD0660w | | Descriptor: | GLYCEROL, PHOSPHATE ION, putative Phosphoglycerate mutase | | Authors: | Wernimont, A.K, Tempel, W, Lam, A, Zhao, Y, Lew, J, Lin, Y.H, Wasney, G, Vedadi, M, Kozieradzki, I, Cossar, D, Schapira, M, Weigelt, J, Arrowsmith, C.H, Bochkarev, A, Edwards, A.M, Hui, R, Pizarro, J, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a new phosphatase from Plasmodium.
Mol.Biochem.Parasitol., 179, 2011
|
|
3E9E
 
 | |
