2FJE
 
 | | adenosine-5-phosphosulfate reductase oxidized state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, adenylylsulfate reductase, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
2FJB
 
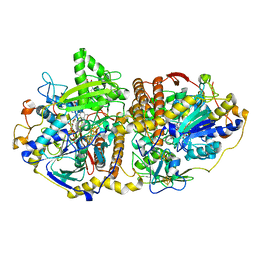 | | Adenosine-5'-phosphosulfate reductase im complex with products | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, ADENOSINE MONOPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
1A6L
 
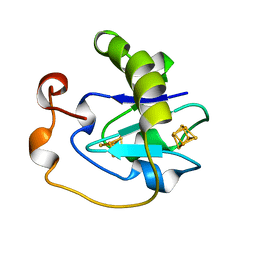 | | T14C MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Gao-Sheridan, H.S, Kemper, M.A, Khayat, R, Armstrong, F.A, Prasad, G.S, Sridhar, V, Stout, C.D, Burgess, B.K. | | Deposit date: | 1998-02-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A T14C variant of Azotobacter vinelandii ferredoxin I undergoes facile [3Fe-4S]0 to [4Fe-4S]2+ conversion in vitro but not in vivo.
J.Biol.Chem., 273, 1998
|
|
7Z83
 
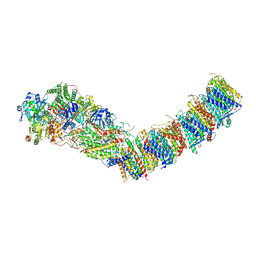 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z84
 
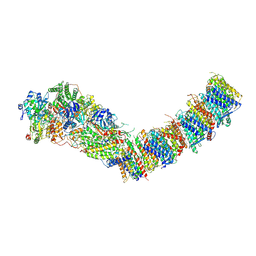 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7S
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z80
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7V
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7R
 
 | | Complex I from E. coli, LMNG-purified, Apo, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z0T
 
 | | Structure of the Escherichia coli formate hydrogenlyase complex (aerobic preparation, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBONMONOXIDE-(DICYANO) IRON, FE (III) ION, ... | | Authors: | Steinhilper, R, Murphy, B.J. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli.
Nat Commun, 13, 2022
|
|
7Z0S
 
 | |
1B0V
 
 | | I40N MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN) | | Authors: | Sridhar, V, Prasad, G.S, Stout, C.D, Chen, K, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Alteration of the reduction potential of the [4Fe-4S](2+/+) cluster of Azotobacter vinelandii ferredoxin I.
J.Biol.Chem., 274, 1999
|
|
1B0P
 
 | | CRYSTAL STRUCTURE OF PYRUVATE-FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chabriere, E, Charon, M.H, Volbeda, A. | | Deposit date: | 1998-11-12 | | Release date: | 1999-04-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.
Nat.Struct.Biol., 6, 1999
|
|
7Z7T
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
1B0T
 
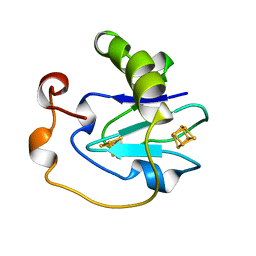 | | D15K/K84D MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN I) | | Authors: | Sridhar, V, Stout, C.D, Chen, K, Kemper, M.A, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the D15K/K84D Mutant of Azotobacter Vinelandii Ferredoxin I
To be Published
|
|
7ZC5
 
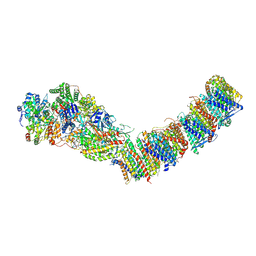 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
1AXQ
 
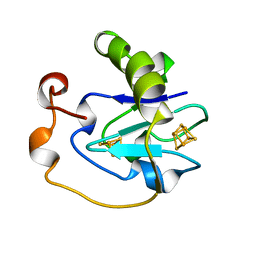 | | FERRICYANIDE OXIDIZED FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of ferricyanide-oxidized [Fe-S] clusters in Azotobacter vinelandii ferredoxin I.
J.Biol.Inorg.Chem., 3, 1998
|
|
3MM5
 
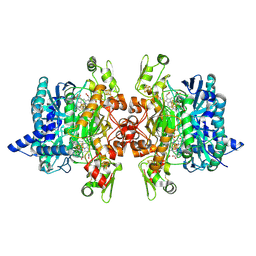 | | Dissimilatory sulfite reductase in complex with the substrate sulfite | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
6ZJN
 
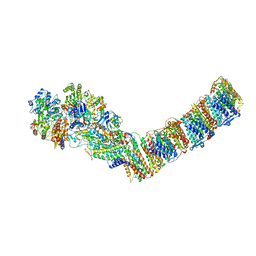 | | Respiratory complex I from Thermus thermophilus, NADH dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZJL
 
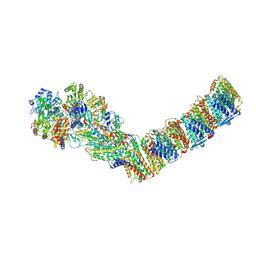 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, major state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
3MM6
 
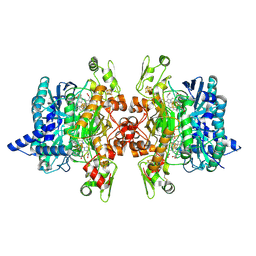 | | Dissimilatory sulfite reductase cyanide complex | | Descriptor: | CYANIDE ION, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3M9S
 
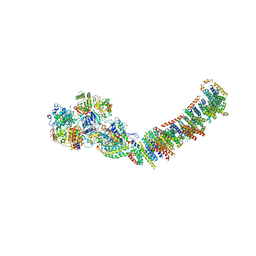 | | Crystal structure of respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Efremov, R.G, Baradaran, R, Sazanov, L.A. | | Deposit date: | 2010-03-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The architecture of respiratory complex I
Nature, 465, 2010
|
|
3OR2
 
 | | Crystal structure of dissimilatory sulfite reductase II (DsrII) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissimilatory Sulfite Reductase, Sulfate Reduction
To be Published
|
|
3MM9
 
 | | Dissimilatory sulfite reductase nitrite complex | | Descriptor: | IRON/SULFUR CLUSTER, NITRITE ION, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MMB
 
 | | Dissimilatory sulfite reductase in complex with the endproduct sulfide | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
