1ADD
 
 | |
8OO6
 
 | | Pol I bound to extended and displaced DNA section - closed conformation | | Descriptor: | DNA polymerase I, Displaced primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1AKZ
 
 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
5Y3B
 
 | |
8EPL
 
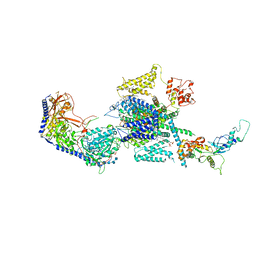 | | Human R-type voltage-gated calcium channel Cav2.3 at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
8OOY
 
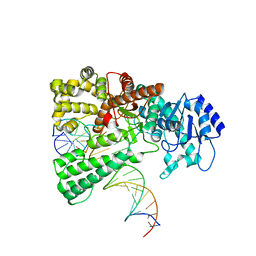 | | Pol I bound to extended and displaced DNA section - open conformation | | Descriptor: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8USS
 
 | | IL17A complexed to Compound 7 | | Descriptor: | 4,5-dichloro-N-[(1S)-1-cyclohexyl-2-{[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]amino}-2-oxoethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, Interleukin-17A | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
8EEB
 
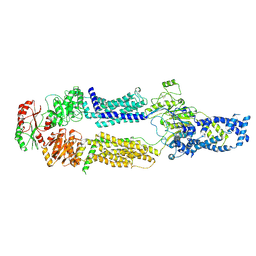 | | Cryo-EM structure of human ABCA7 in Digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
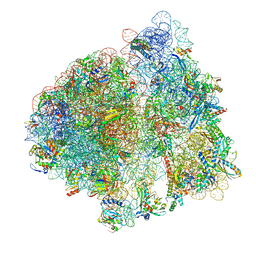
8EKC
 
 | |
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8EOP
 
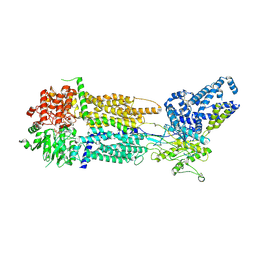 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
5YSY
 
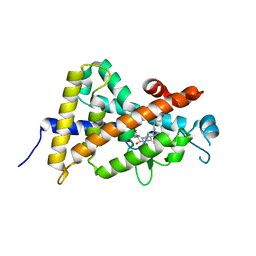 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2R,3R)-5-[(E)-2-{(1R,3aS,7aR)-1-[(R)-6-hydroxy-6-methylheptan-2-yl]-7a-methyl-2,3,3a,6,7,7a-hexahydro-1H-inden-4-yl}vinyl]-2-(3-hydroxypropyl)cyclohex-4-ene-1,3-diol | | Descriptor: | (1R,2R,3R)-5-[(E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-1,2,3,3a,6,7-hexahydroinden-4-yl]e thenyl]-2-(3-oxidanylpropyl)cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Takimoto-Kamimura, M, Kakuda, S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of 2-substitution on 14-epi-19-nortachysterol-mediated biological events: based on synthesis and X-ray co-crystallographic analysis with the human vitamin D receptor.
Org. Biomol. Chem., 16, 2018
|
|
1AL8
 
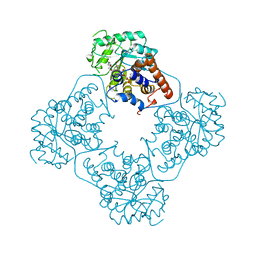 | |
1AM2
 
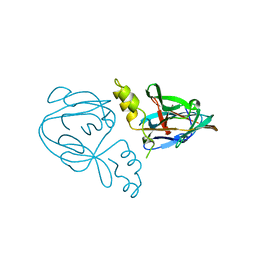 | |
5YT2
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R)-5-[(E)-2-{(1R,3aS,7aR)-1-[(R)-6-hydroxy-6-methylheptan-2-yl]-7a-methyl-2,3,3a,6,7,7a-hexahydro-1H-inden-4-yl}vinyl]-2-(3-hydroxypropyl)cyclohex-4-ene-1,3-diol | | Descriptor: | (1R,2S,3R)-5-[(E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-1,2,3,3a,6,7-hexahydroinden-4-yl]ethenyl]-2-(3-oxidanylpropyl)cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Takimoto-Kamimura, M, Kakuda, S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of 2-substitution on 14-epi-19-nortachysterol-mediated biological events: based on synthesis and X-ray co-crystallographic analysis with the human vitamin D receptor.
Org. Biomol. Chem., 16, 2018
|
|
8V85
 
 | |
8V83
 
 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8EE6
 
 | | Cryo-EM Structure of human ABCA7 in PE/Ch nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EDW
 
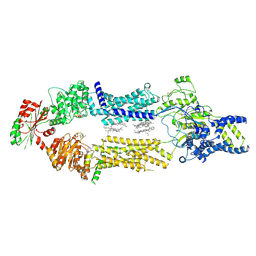 | | Cryo-EM Structure of human ABCA7 in BPL/Ch Nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
5Y5G
 
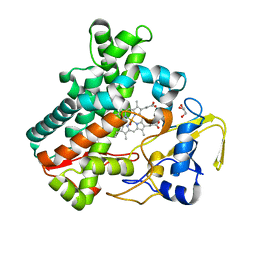 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
8P3X
 
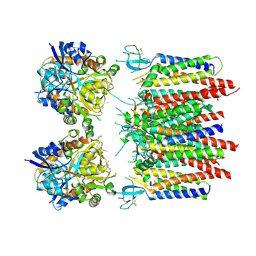 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
5Y5K
 
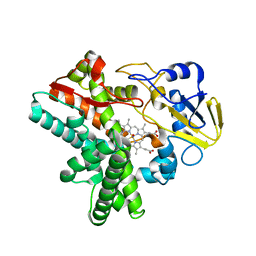 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
8P3S
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
1AL7
 
 | |
8OFT
 
 | |
