5RVZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DHTKD1 in complex with Z1929757385 | | Descriptor: | (3S)-1-(3-fluoropyridin-2-yl)-4,4-dimethylpyrrolidin-3-ol, MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RWW
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2241115980 | | Descriptor: | 1-[(furan-2-yl)methyl]-4-(methylsulfonyl)piperazine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RXF
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2074076908 | | Descriptor: | 1-(5-azaspiro[2.5]octan-5-yl)-2-(difluoromethoxy)ethan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RX8
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434884 | | Descriptor: | 1-[4-(3-phenylpropyl)piperazin-1-yl]ethan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RXX
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1275599911 | | Descriptor: | 1-(2-azaspiro[5.6]dodecan-2-yl)ethan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RXQ
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z26548083 | | Descriptor: | DIMETHYL SULFOXIDE, N-(3-cyanophenyl)-2-methylpropanamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYD
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z955369596 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(5-bromo-2-methoxyphenyl)methyl]acetamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RY5
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z166605480 | | Descriptor: | (1S)-1-(4-fluorophenyl)-N-methylethan-1-amine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYT
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z54226006 | | Descriptor: | (2,6-difluorophenyl)(pyrrolidin-1-yl)methanone, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RYH
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1212984951 | | Descriptor: | (3S)-3-(methylamino)-1-phenylpiperidin-2-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RZ5
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434851 | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-fluorophenyl)methyl]amino}methyl)benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZ6
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434944 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZG
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z133729708 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZM
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434815 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(dimethylamino)methyl]benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZW
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2574937229 | | Descriptor: | (S)-1-(4-methoxyphenyl)-1-phenylmethanamine, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RPR
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo15 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROB
 
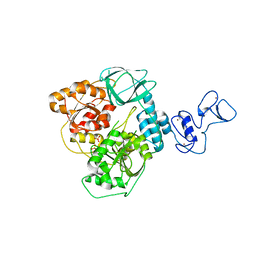 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 helicase | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5ROJ
 
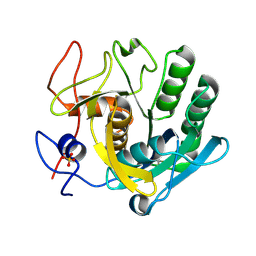 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo59 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROE
 
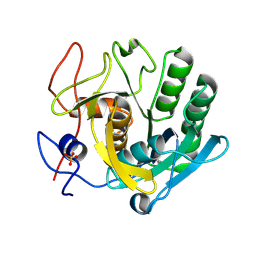 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo6 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROQ
 
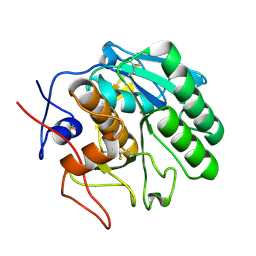 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen A12a | | Descriptor: | (2R)-(3-chlorophenyl)(hydroxy)ethanoic acid, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROF
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen C11a | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPL
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen C8a | | Descriptor: | (4-methoxycarbonylphenyl)methylazanium, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROH
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo61 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROI
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo30 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPS
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo60 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
