8BAC
 
 | | Crystal structure of human heparanase in complex with competitive inhibitor GD05 | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of Uronic Acid 1-Azasugars as Putative Inhibitors of alpha-Iduronidase, beta-Glucuronidase and Heparanase.
Chembiochem, 24, 2023
|
|
7ADY
 
 | | CO-removed state of the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8THM
 
 | |
1FMC
 
 | | 7-ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEX WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1996-04-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
8BAJ
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
8PZ8
 
 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 54 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
7ADR
 
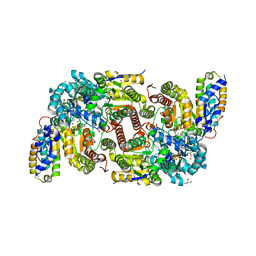 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1FNB
 
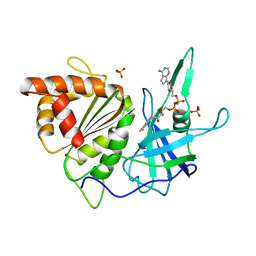 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FNC
 
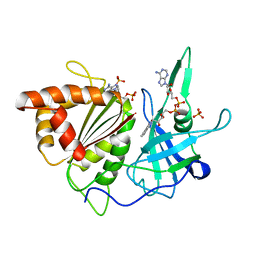 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FERREDOXIN-NADP+ REDUCTASE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FND
 
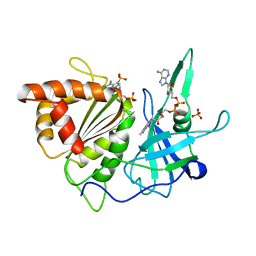 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
5C5W
 
 | | 1.25 A resolution structure of an RNA 20-mer | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*AP*GP*UP*UP*CP*AP*AP*UP*UP*CP*UP*AP*GP*CP*G)-3') | | Authors: | Stewart, M, Valkov, E. | | Deposit date: | 2015-06-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8ZJ5
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate. | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate.
To Be Published
|
|
8ZJ0
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3-Hydroxybenzoate. | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4-hydroxythreonine-4-phosphate dehydrogenase, CHLORIDE ION, ... | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3-Hydroxybenzoate.
To Be Published
|
|
8ZJ7
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate and NAD. | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxythreonine-4-phosphate dehydrogenase, CHLORIDE ION, ... | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate and NAD.
To Be Published
|
|
8RFC
 
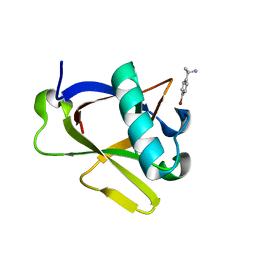 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL1 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-bromophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFD
 
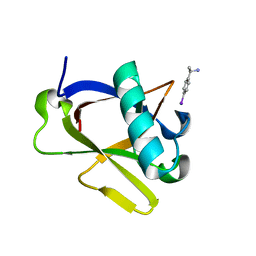 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9DO4
 
 | |
9DR6
 
 | |
8R2H
 
 | | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Plasmodium falciparum | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gawriljuk, V.O, Godoy, A.S, Oerlemans, R, Groves, M.R. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase DXPS from Plasmodium falciparum reveals a distinct N-terminal domain.
Nat Commun, 15, 2024
|
|
8WUE
 
 | |
5G2O
 
 | | Yersinia pestis FabV variant T276A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-11 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
6T3E
 
 | | Structure of Thermococcus litoralis Delta(1)-pyrroline-2-carboxylate reductase in complex with NADH and L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DELTA1-pyrroline-2-carboxylate reductase, PROLINE | | Authors: | Ferraris, D.M, Miggiano, R, Ferrario, E, Rizzi, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermococcus litoralis Delta1-pyrroline-2-carboxylate reductase in complex with NADH and L-proline.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7QLG
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT L259V COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE (III) ION, GLYCEROL, ... | | Authors: | Sridhar, S, Kiema, T.R, Widertsen, M, Wierenga, R.K. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
8RNP
 
 | |
9DSN
 
 | | D306A Mutant of M.tuberculosis MenD (SEPHCHC Synthase) | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Ho, N.A.T, Given, F.M, Allison, T.A, Bulloch, E.M.M, Jiao, W. | | Deposit date: | 2024-09-28 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Apparent Reversal of Allosteric Response in Mycobacterium tuberculosis MenD Reveals Links to Half-of-Sites Reactivity.
Chembiochem, 26, 2025
|
|
