5KND
 
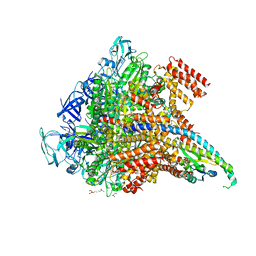 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
5LQY
 
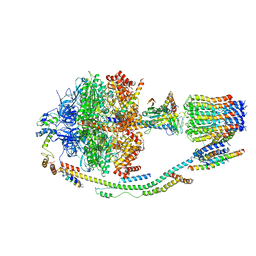 | | Structure of F-ATPase from Pichia angusta, in state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LQX
 
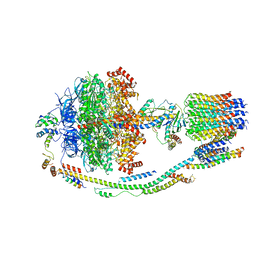 | | Structure of F-ATPase from Pichia angusta, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase AAP1 subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5IK2
 
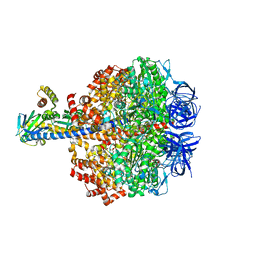 | | Caldalaklibacillus thermarum F1-ATPase (epsilon mutant) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1OHH
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE complexed with the inhibitor protein IF1 | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Cabezon, E, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Bovine F1-ATPase in Complex with its Regulatory Protein If1
Nat.Struct.Biol., 10, 2003
|
|
6B8H
 
 | | Mosaic model of yeast mitochondrial ATP synthase monomer | | Descriptor: | AATP synthase subunit g, ATP synthase catalytic sector F1 epsilon subunit, ATP synthase protein 8, ... | | Authors: | Guo, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2017-10-07 | | Release date: | 2018-01-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic model for the dimeric FO region of mitochondrial ATP synthase.
Science, 358, 2017
|
|
6F5D
 
 | | Trypanosoma brucei F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma subunit, ATP synthase subunit alpha, ... | | Authors: | Montgomery, M.G, Gahura, O, Leslie, A.G.W, Zikova, A, Walker, J.E. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ATP synthase fromTrypanosoma bruceihas an elaborated canonical F1-domain and conventional catalytic sites.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1W0K
 
 | | ADP inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
1W0J
 
 | | Beryllium fluoride inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
6FKH
 
 | | Chloroplast F1Fo conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
1XPR
 
 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-formylbicyclomycin (FB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-FORMYLBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
6FKF
 
 | | Chloroplast F1Fo conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
5LQZ
 
 | | Structure of F-ATPase from Pichia angusta, state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JJI
 
 | | Rho transcription termination factor bound to rU7 and 6 ADP-BeF3 molecules | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JJL
 
 | | Rho transcription termination factor bound to rU8 and 5 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1PV4
 
 | |
1PVO
 
 | |
6FOC
 
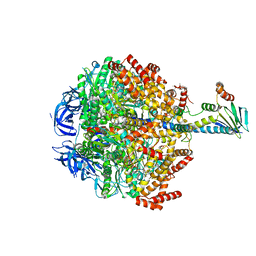 | | F1-ATPase from Mycobacterium smegmatis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, T, Montgomery, M.G, Leslie, A.G.W, Cook, G.M, Walker, J.E. | | Deposit date: | 2018-02-06 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structure of the catalytic domain of the ATP synthase fromMycobacterium smegmatisis a target for developing antitubercular drugs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1XPU
 
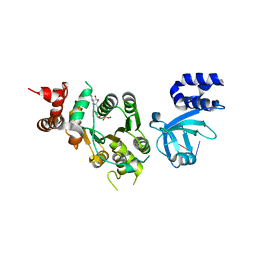 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-(3-FORMYLPHENYLSULFANYL)-DIHYDROBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1XPO
 
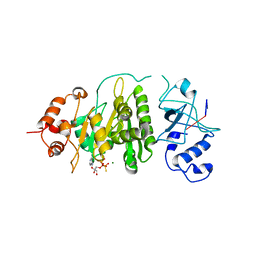 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', BICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2005-02-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
5HKK
 
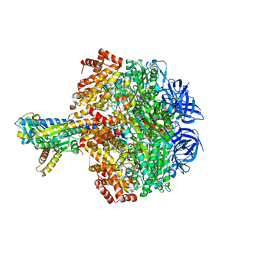 | | Caldalaklibacillus thermarum F1-ATPase (wild type) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6CP6
 
 | | Monomer yeast ATP synthase (F1Fo) reconstituted in nanodisc. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
5JJK
 
 | | Rho transcription termination factor bound to rA7 and 6 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6FKI
 
 | | Chloroplast F1Fo conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
