1L0R
 
 | |
1RMG
 
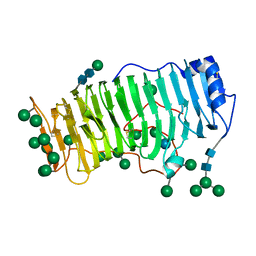 | | RHAMNOGALACTURONASE A FROM ASPERGILLUS ACULEATUS | | Descriptor: | RHAMNOGALACTURONASE A, alpha-D-glucopyranose, alpha-D-mannopyranose, ... | | Authors: | Petersen, T.N, Kauppinen, S, Larsen, S. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix.
Structure, 5, 1997
|
|
1W2T
 
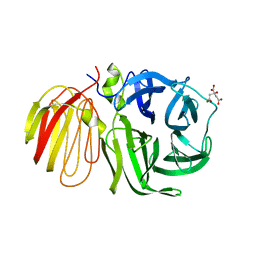 | | beta-fructosidase from Thermotoga maritima in complex with raffinose | | Descriptor: | BETA FRUCTOSIDASE, CITRIC ACID, SULFATE ION, ... | | Authors: | Alberto, F, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-07-08 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of Inactivated Thermotoga Maritima Invertase in Complex with the Trisaccharide Substrate Raffinose.
Biochem.J., 395, 2006
|
|
282D
 
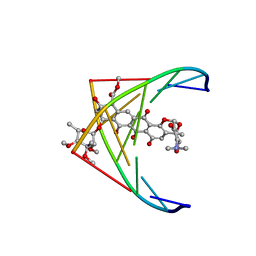 | | A CONTINOUS TRANSITION FROM A-DNA TO B-DNA IN THE 1:1 COMPLEX BETWEEN NOGALAMYCIN AND THE HEXAMER DCCCGGG | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), NOGALAMYCIN | | Authors: | Cruse, W, Saludjian, P, Leroux, Y, Leger, Y, El Manouni, D, Prange, T. | | Deposit date: | 1996-08-26 | | Release date: | 1996-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A continuous transition from A-DNA to B-DNA in the 1:1 complex between nogalamycin and the hexamer dCCCGGG.
J.Biol.Chem., 271, 1996
|
|
3PE7
 
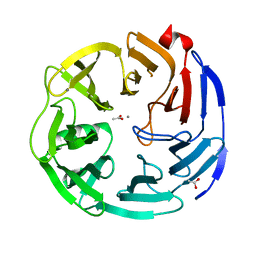 | | Oligogalacturonate lyase in complex with manganese | | Descriptor: | ACETATE ION, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Abbott, D.W, Gilbert, H.J, Boraston, A.B. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The active site of oligogalacturonate lyase provides unique insights into cytoplasmic oligogalacturonate beta-elimination.
J.Biol.Chem., 285, 2010
|
|
2TMG
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT S128R, T158E, N117R, S160E | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, van der Oost, J, de Vos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface.
J.Mol.Biol., 289, 1999
|
|
4DDX
 
 | |
4DDU
 
 | | Thermotoga maritima reverse gyrase, C2 FORM 1 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDT
 
 | | Thermotoga maritima reverse gyrase, C2 FORM 2 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDW
 
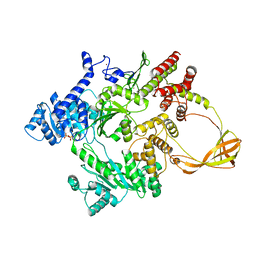 | |
4DDV
 
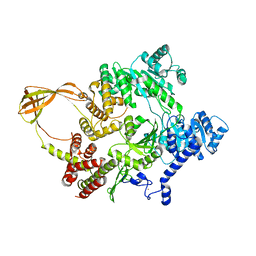 | |
7OU8
 
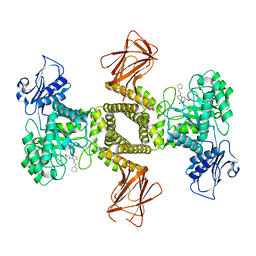 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
1XJM
 
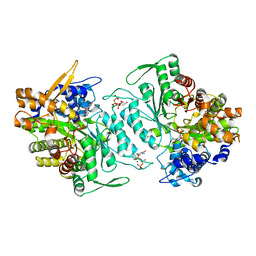 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dTTP complex | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE, ribonucleotide reductase, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XJK
 
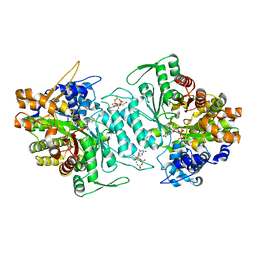 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dGTP-ADP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XJJ
 
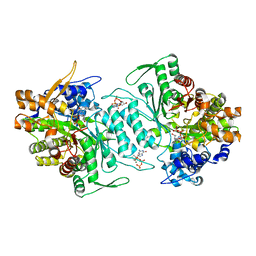 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dGTP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ribonucleotide reductase, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2005
|
|
1XJG
 
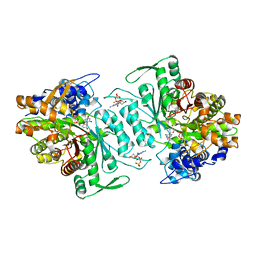 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dATP-UDP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XJN
 
 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dATP-CDP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XJF
 
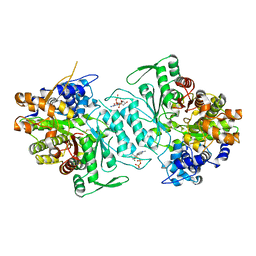 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, ribonucleotide reductase, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1ROF
 
 | | NMR STUDY OF 4FE-4S FERREDOXIN OF THERMATOGA MARITIMA | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Roesch, P, Sticht, H, Wildegger, G, Bentrop, D, Darimont, B, Sterner, R. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized Thermotoga maritima 1[Fe4-S4] ferredoxin.
Eur.J.Biochem., 237, 1996
|
|
1XJE
 
 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dTTP-GDP complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3A06
 
 | | Crystal structure of DXR from Thermooga maritia, in complex with fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Takenoya, M, Ohtaki, A, Noguchi, K, Sasaki, Y, Ohsawa, K, Yohda, M, Yajima, S. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding
J.Struct.Biol., 170, 2010
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
1A5Z
 
 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
3TMY
 
 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
3T8Y
 
 | |
