1AR9
 
 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AS3
 
 | | GDP BOUND G42V GIA1 | | Descriptor: | GIA1, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1AS0
 
 | | GTP-GAMMA-S BOUND G42V GIA1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GIA1, MAGNESIUM ION, ... | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1AS2
 
 | | GDP+PI BOUND G42V GIA1 | | Descriptor: | GIA1, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1ASK
 
 | | NUCLEAR TRANSPORT FACTOR 2 (NTF2) H66A MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Mccoy, A.J, Stewart, M.J. | | Deposit date: | 1997-08-11 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran.
J.Mol.Biol., 272, 1997
|
|
1ASS
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, SODIUM ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1ASX
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1AR7
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR8
 
 | | P1/MAHONEY POLIOVIRUS, MUTANT P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1ASJ
 
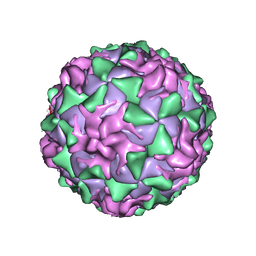 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR6
 
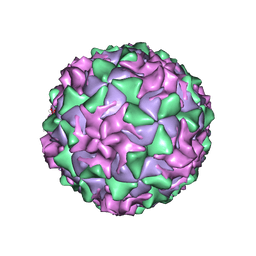 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
5GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
1AS4
 
 | | CLEAVED ANTICHYMOTRYPSIN A349R | | Descriptor: | ACETATE ION, ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1UUU
 
 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1IRB
 
 | | CARBOXYLIC ESTER HYDROLASE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-08-13 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phospholipase A2 engineering. Deletion of the C-terminus segment changes substrate specificity and uncouples calcium and substrate binding at the zwitterionic interface.
Biochemistry, 35, 1996
|
|
1AS8
 
 | |
1AS7
 
 | |
1AS6
 
 | |
2IZA
 
 | | APOSTREPTAVIDIN PH 2.00 I4122 STRUCTURE | | Descriptor: | FORMIC ACID, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZE
 
 | | APOSTREPTAVIDIN PH 3.08 I222 COMPLEX | | Descriptor: | AMMONIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZJ
 
 | | STREPTAVIDIN-BIOTIN PH 3.50 I4122 STRUCTURE | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZD
 
 | | APOSTREPTAVIDIN pH 3.0 I222 COMPLEX | | Descriptor: | AMMONIUM ION, CHLORIDE ION, IODIDE ION, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZB
 
 | | APOSTREPTAVIDIN PH 3.12 I4122 STRUCTURE | | Descriptor: | FORMIC ACID, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZL
 
 | | STREPTAVIDIN-2-IMINOBIOTIN PH 7.3 I222 COMPLEX | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZK
 
 | | STREPTAVIDIN-GLYCOLURIL PH 2.58 I4122 COMPLEX | | Descriptor: | ACETATE ION, GLYCOLURIL, STREPTAVIDIN, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
