1C0Q
 
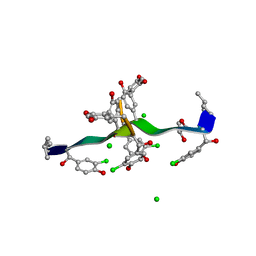 | | COMPLEX OF VANCOMYCIN WITH 2-ACETOXY-D-PROPANOIC ACID | | Descriptor: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin binding to low-affinity ligands: delineating a minimum set of interactions necessary for high-affinity binding.
J.Med.Chem., 42, 1999
|
|
182D
 
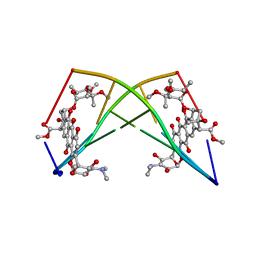 | | DNA-NOGALAMYCIN INTERACTIONS: THE CRYSTAL STRUCTURE OF D(TGATCA) COMPLEXED WITH NOGALAMYCIN | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Smith, C.K, Davies, G.J, Dodson, E.J, Moore, M.H. | | Deposit date: | 1994-07-28 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA-nogalamycin interactions: the crystal structure of d(TGATCA) complexed with nogalamycin.
Biochemistry, 34, 1995
|
|
5YIO
 
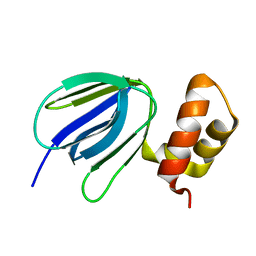 | | NMR solution structure of subunit epsilon of the Mycobacterium tuberculosis F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Ragunathan, P, Sundararaman, L, Nartey, W, Manimekalai, M.S.S, Bogdanovic, N, Gruber, G. | | Deposit date: | 2017-10-06 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of Mycobacterium tuberculosis F-ATP synthase subunit epsilon provides new insight into energy coupling inside the rotary engine.
FEBS J., 285, 2018
|
|
8PNL
 
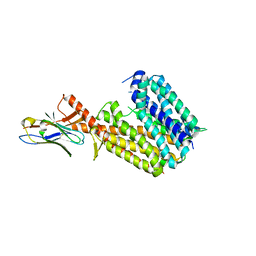 | | Outward-open conformation of a Major Facilitator Superfamily (MFS) transporter MHAS2168, a homologue of Rv1410 from M. tuberculosis, in complex with an alpaca nanobody | | Descriptor: | Nb_H2, Putative triacylglyceride transporter | | Authors: | Remm, S, Schoeppe, J, Hutter, C.A.J, Gonda, I, Seeger, M.A. | | Deposit date: | 2023-06-30 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for triacylglyceride extraction from mycobacterial inner membrane by MFS transporter Rv1410.
Nat Commun, 14, 2023
|
|
9IMR
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase Rv0562 from Mycobacterium tuberculosis in complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, BICINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Q, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insight of a bi-functional isoprenyl diphosphate synthase Rv0562 from Mycobacterium tuberculosis.
Int.J.Biol.Macromol., 2025
|
|
9IMQ
 
 | | Crystal structure of a C45 isoprenyl diphosphate synthase, Rv0562 from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, Nonaprenyl diphosphate synthase | | Authors: | Wang, Q, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural insight of a bi-functional isoprenyl diphosphate synthase Rv0562 from Mycobacterium tuberculosis.
Int.J.Biol.Macromol., 2025
|
|
9IMS
 
 | |
9KUQ
 
 | |
6AX8
 
 | | Mycobacterium tuberculosis methionyl-tRNA synthetase in complex with methionyl-adenylate | | Descriptor: | Methionine-tRNA ligase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] (2S)-2-azanyl-4-methylsulfanyl-butanoate | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2017-09-06 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the drug target Mycobacterium tuberculosis methionyl-tRNA synthetase in complex with a catalytic intermediate.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8S9D
 
 | |
8S97
 
 | |
6FUX
 
 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 in complex with ADP and streptomycin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6A2S
 
 | | Mycobacterium tuberculosis LexA C-domain S160A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A2Q
 
 | | Mycobacterium tuberculosis LexA C-domain I | | Descriptor: | GLYCEROL, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A2R
 
 | | Mycobacterium tuberculosis LexA C-domain II | | Descriptor: | DI(HYDROXYETHYL)ETHER, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6ACA
 
 | | Crystal structure of Mycobacterium tuberculosis Mfd at 3.6 A resolution | | Descriptor: | Mycobacterium tuberculosis Mfd | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6A2T
 
 | | Mycobacterium tuberculosis LexA C-domain K197A | | Descriptor: | ACRYLIC ACID, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6BS8
 
 | | The class 3 DnaB intein from Mycobacterium smegmatis | | Descriptor: | Replicative DNA helicase | | Authors: | Li, Z, Kelley, D.S, Banavali, N, Belfort, M, Li, H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mycobacterial DnaB helicase intein as oxidative stress sensor.
Nat Commun, 9, 2018
|
|
3RTK
 
 | | Crystal structure of Cpn60.2 from Mycobacterium tuberculosis at 2.8A | | Descriptor: | 60 kDa chaperonin 2, MAGNESIUM ION | | Authors: | Shahar, A, Melamed-Frank, M, Kashi, Y, Adir, N. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The dimeric structure of the Cpn60.2 chaperonin of Mycobacterium tuberculosis at 2.8 A reveals possible modes of function.
J.Mol.Biol., 412, 2011
|
|
8G82
 
 | | Vancomycin bound to D-Ala-D-Ser | | Descriptor: | BORIC ACID, D-Ala-D-Ser, DIMETHYL SULFOXIDE, ... | | Authors: | Loll, P.J, Park, J.H. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of vancomycin bound to the resistance determinant D-alanine-D-serine.
Iucrj, 11, 2024
|
|
3QB9
 
 | |
1CHK
 
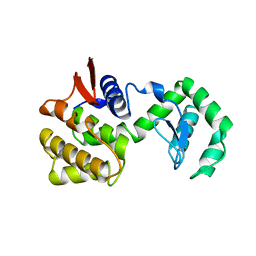 | |
1D17
 
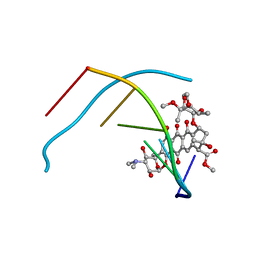 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
7NM8
 
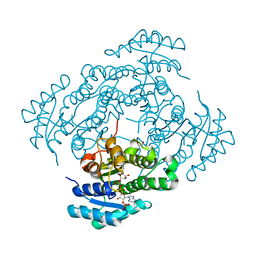 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM, in complex with NADPH | | Descriptor: | Antimycin pathway standalone ketoreductase, AntM, GLYCEROL, ... | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
7NM7
 
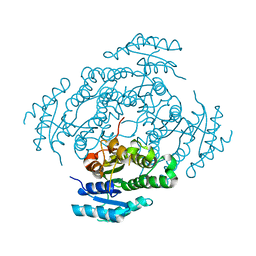 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM | | Descriptor: | Antimycin pathway standalone ketoreductase enzyme, AntM | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
