1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1W5W
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,4-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1R,2S)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
7WAB
 
 | | Crystal structure of the prolyl endoprotease, PEP, from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPASS (Complex proteins associated with Set1p) component shg1 family protein, ... | | Authors: | Miyazono, K, Kubota, K, Takahashi, K, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate recognition mechanism of the prolyl endoprotease PEP from Aspergillus niger.
Biochem.Biophys.Res.Commun., 591, 2022
|
|
6G9V
 
 | | Crystal structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase(AfUAP1) in complex with UDPGlcNAc, pyrophosphate and Mg2+ | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, UDP-N-acetylglucosamine pyrophosphorylase, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
6G9W
 
 | | Crystal Structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase in complex with UTP | | Descriptor: | UDP-N-acetylglucosamine pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
6GUO
 
 | | Siderophore hydrolase EstA from Aspergillus nidulans | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative siderophore-degrading esterase (Eurofung), ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GUI
 
 | | Siderophore hydrolase EstB mutant H267N from Aspergillus fumigatus | | Descriptor: | GLYCEROL, Siderophore esterase IroE-like, putative | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GSG
 
 | | Crystal structure of Aspergillus oryzae catechol oxidase complexed with resorcinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Catechol oxidase, ... | | Authors: | Penttinen, L, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Unraveling Substrate Specificity and Catalytic Promiscuity of Aspergillus oryzae Catechol Oxidase.
Chembiochem, 19, 2018
|
|
6GUD
 
 | | Siderophore hydrolase EstB from Aspergillus fumigatus | | Descriptor: | CARBONATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GUP
 
 | | Siderophore hydrolase EstB from Aspergillus fumigatus | | Descriptor: | GLYCEROL, Siderophore biosynthesis lipase/esterase, putative | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GUR
 
 | | Siderophore hydrolase EstB from Aspergillus fumigatus in complex with TAFC | | Descriptor: | (~{Z})-5-[(1~{S},2~{S})-2-acetamido-1-oxidanyl-5-[oxidanyl(propanoyl)amino]pentoxy]-~{N},3-dimethyl-~{N}-oxidanyl-pent-2-enamide, CARBONATE ION, FE (III) ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7A1A
 
 | |
7A19
 
 | |
6GUL
 
 | | Siderophore hydrolase EstB mutant E211Q from Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GUG
 
 | | Siderophore hydrolase EstB mutant S148A from Aspergillus fumigatus | | Descriptor: | GLYCEROL, SULFATE ION, Siderophore esterase IroE-like, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6I1A
 
 | | Crystal structure of rutinosidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, rutinosidase | | Authors: | Pachl, P, Rezacova, P, Kapesova, J. | | Deposit date: | 2018-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rutinosidase from Aspergillus niger: crystal structure and insight into the enzymatic activity.
Febs J., 287, 2020
|
|
1RNR
 
 | |
1UET
 
 | |
1UEU
 
 | |
7MO6
 
 | |
7A3M
 
 | |
8HMM
 
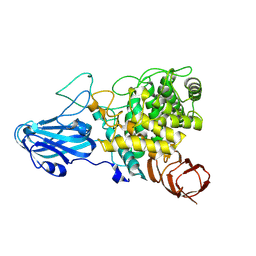 | | Crystal structure of AoRhaA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bac_rhamnosid6H domain-containing protein | | Authors: | Makabe, K, Koseki, T. | | Deposit date: | 2022-12-05 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aspergillus oryzae alpha-l-rhamnosidase: Crystal structure and insight into the substrate specificity.
Proteins, 92, 2024
|
|
6ZLO
 
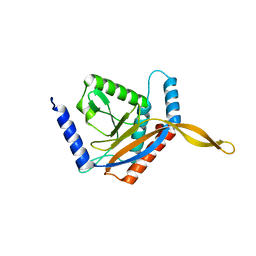 | | E2 core of the fungal Pyruvate dehydrogenase complex with asymmetric interior PX30 component | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Forsberg, B.O, Howard, R.J, Aibara, S, Mortesaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
6ZLM
 
 | | Dihydrolipoyllysine-residue acetyltransferase component of fungal pyruvate dehydrogenase complex with protein X bound | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, Pyruvate dehydrogenase X component | | Authors: | Forsberg, B.O, Aibara, S, Howard, R.J, Mortezaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
1FI1
 
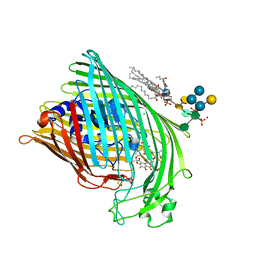 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
