1DOY
 
 | | 1H AND 15N SEQUENTIAL ASSIGNMENT, SECONDARY STRUCTURE AND TERTIARY FOLD OF [2FE-2S] FERREDOXIN FROM SYNECHOCYSTIS SP. PCC 6803 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S] | | Authors: | Lelong, C, Setif, P, Bottin, H, Andre, F, Neumann, J.M. | | Deposit date: | 1995-09-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803.
Biochemistry, 34, 1995
|
|
5M2G
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 2,4,6-tribromophenol | | Descriptor: | 2,4,6-TRIBROMOPHENOL, BENZAMIDINE, GLYCEROL, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-10-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
1DSB
 
 | |
1DBJ
 
 | |
1DCL
 
 | |
1DKJ
 
 | | BOBWHITE QUAIL LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
5M30
 
 | | Structure of TssK from T6SS EAEC in complex with nanobody nb18 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Type VI secretion protein | | Authors: | Nguyen, V.S, Cambillau, C, Spinelli, C, Desmyter, A, Legrand, P, Cascales, E. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
1MH0
 
 | | Crystal structure of the anticoagulant slow form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pineda, A.O, Savvides, S, Waksman, G, Di Cera, E. | | Deposit date: | 2002-08-18 | | Release date: | 2002-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the anticoagulant slow form of thrombin
J.Biol.Chem., 277, 2002
|
|
1DIN
 
 | |
5M3O
 
 | | HTRA2 A141S mutant structure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Serine protease HTRA2, mitochondrial | | Authors: | Merski, M, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
1DWC
 
 | |
5M3U
 
 | | The X-ray structure of human V216F phosphoglycerate kinase 1 mutant | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
1DX1
 
 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5M44
 
 | | Complex structure of human protein kinase CK2 catalytic subunit with a thieno[2,3-d]pyrimidin inhibitor crystallized under high-salt conditions | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
1DSU
 
 | |
5LFO
 
 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
1DVF
 
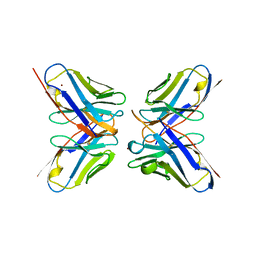 | | IDIOTOPIC ANTIBODY D1.3 FV FRAGMENT-ANTIIDIOTOPIC ANTIBODY E5.2 FV FRAGMENT COMPLEX | | Descriptor: | FV D1.3, FV E5.2, ZINC ION | | Authors: | Braden, B.C, Fields, B.A, Ysern, X, Dall'Acqua, W, Goldbaum, F.A, Poljak, R.J, Mariuzza, R.A. | | Deposit date: | 1996-04-13 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an Fv-Fv idiotope-anti-idiotope complex at 1.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
1DWD
 
 | |
1DYP
 
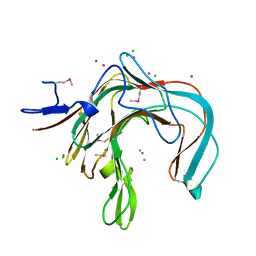 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
1DWM
 
 | |
5LCN
 
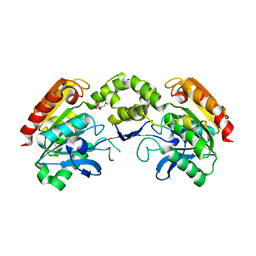 | |
1E1J
 
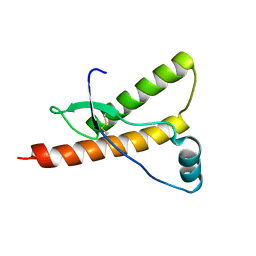 | | Human prion protein variant M166V | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-09 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DX0
 
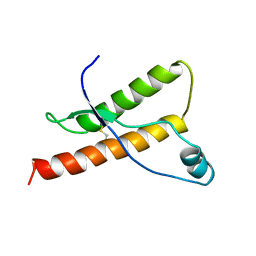 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5LD0
 
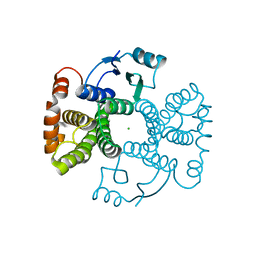 | | Chimeric GST | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1,Glutathione S-transferase alpha-2,Glutathione S-transferase A1 | | Authors: | Axarli, A, Muleta, A.W, Chronopoulou, E.G, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution of glutathione transferases towards a selective glutathione-binding site and improved oxidative stability.
Biochim. Biophys. Acta, 1861, 2017
|
|
1DWY
 
 | | Bovine prion protein fragment 121-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
