1EMR
 
 | | CRYSTAL STRUCTURE OF HUMAN LEUKEMIA INHIBITORY FACTOR (LIF) | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Heath, J.K, Hawkins, N, Samal, B, Jones, E.Y, Betzel, C. | | Deposit date: | 2000-03-17 | | Release date: | 2001-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Species Variation in Receptor Binding Site Revealed by the Medium Resolution X-ray Structure of Human Leukemia Inhibitory Factor
to be published
|
|
1EMS
 
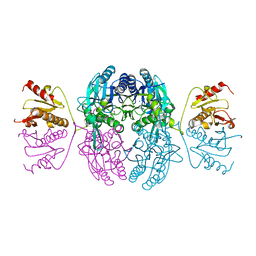 | | CRYSTAL STRUCTURE OF THE C. ELEGANS NITFHIT PROTEIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHYL MERCURY ION, NIT-FRAGILE HISTIDINE TRIAD FUSION PROTEIN, ... | | Authors: | Pace, H.C, Hodawadekar, S.C, Draganescu, A, Huang, J, Bieganowski, P, Pekarsky, Y, Croce, C.M, Brenner, C. | | Deposit date: | 2000-03-17 | | Release date: | 2000-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.
Curr.Biol., 10, 2000
|
|
1EMT
 
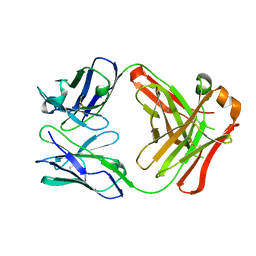 | | FAB ANTIBODY FRAGMENT OF AN C60 ANTIFULLERENE ANTIBODY | | Descriptor: | IGG ANTIBODY (HEAVY CHAIN), IGG ANTIBODY (LIGHT CHAIN) | | Authors: | Braden, B.C, Goldbaum, F.A, Chen, B.-X, Erlanger, B.F, Kirschner, A.N, Wilson, S.R. | | Deposit date: | 2000-03-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystal structure of an anti-Buckminsterfullerene antibody fab fragment: biomolecular recognition of C(60).
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EMU
 
 | |
1EMV
 
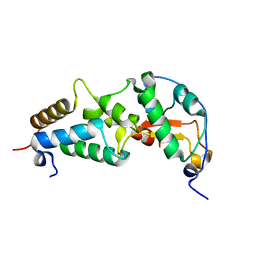 | | CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS) | | Descriptor: | COLICIN E9, IMMUNITY PROTEIN IM9, PHOSPHATE ION | | Authors: | Kuhlmann, U.C, Pommer, A.J, Moore, G.M, James, R, Kleanthous, C. | | Deposit date: | 2000-03-20 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes.
J.Mol.Biol., 301, 2000
|
|
1EMW
 
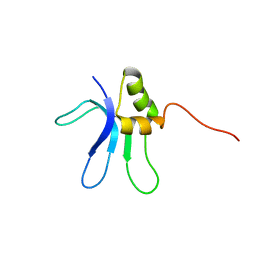 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S16 FROM THERMUS THERMOPHILUS | | Descriptor: | S16 RIBOSOMAL PROTEIN | | Authors: | Allard, P, Rak, A.V, Wimberly, B.T, Clemons Jr, W.M, Kalinin, A, Helgstrand, M, Garber, M.B, Ramakrishnan, V, Hard, T. | | Deposit date: | 2000-03-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Another piece of the ribosome: solution structure of S16 and its location in the 30S subunit.
Structure Fold.Des., 8, 2000
|
|
1EMX
 
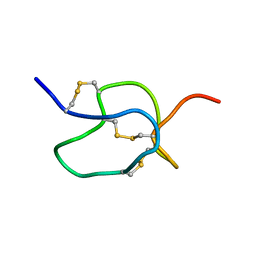 | | SOLUTION STRUCTURE OF HPTX2, A TOXIN FROM HETEROPODA VENATORIA SPIDER VENOM THAT BLOCKS KV4.2 POTASSIUM CHANNEL | | Descriptor: | HETEROPODATOXIN 2 | | Authors: | Bernard, C, Legros, C, Ferrat, G, Bishoff, U, Marquardt, A, Pongs, O, Darbon, H. | | Deposit date: | 2000-03-20 | | Release date: | 2001-01-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hpTX2, a toxin from Heteropoda venatoria spider that blocks Kv4.2 potassium channel.
Protein Sci., 9, 2000
|
|
1EMY
 
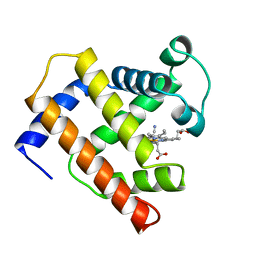 | |
1EMZ
 
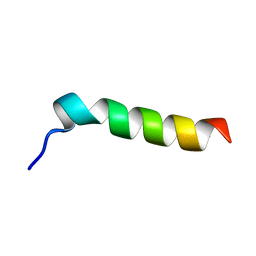 | | SOLUTION STRUCTURE OF FRAGMENT (350-370) OF THE TRANSMEMBRANE DOMAIN OF HEPATITIS C ENVELOPE GLYCOPROTEIN E1 | | Descriptor: | ENVELOPE GLYCOPROTEIN E1 | | Authors: | Op De Beeck, A, Montserret, R, Duvet, S, Cocquerel, L, Cacan, R, Barberot, B, Le Maire, M, Penin, F, Dubuisson, J. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization.
J.Biol.Chem., 275, 2000
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
1EN2
 
 | | UDA TETRASACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-20 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1EN3
 
 | |
1EN4
 
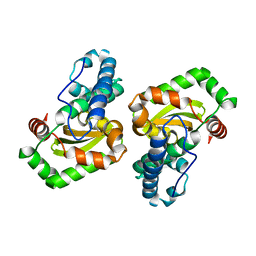 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146H MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN5
 
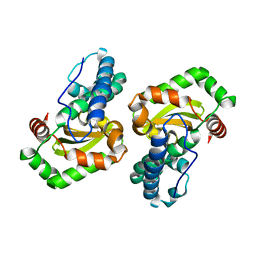 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Y34F MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN6
 
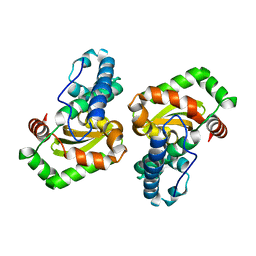 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146L MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN7
 
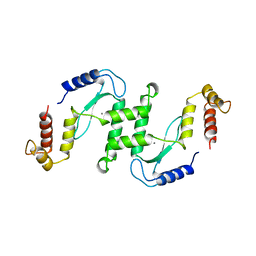 | | ENDONUCLEASE VII (ENDOVII) FROM PHAGE T4 | | Descriptor: | CALCIUM ION, RECOMBINATION ENDONUCLEASE VII, ZINC ION | | Authors: | Raaijmakers, H, Vix, O, Toro, I, Suck, D. | | Deposit date: | 1999-02-07 | | Release date: | 2000-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture.
EMBO J., 18, 1999
|
|
1EN8
 
 | |
1EN9
 
 | |
1ENA
 
 | |
1ENC
 
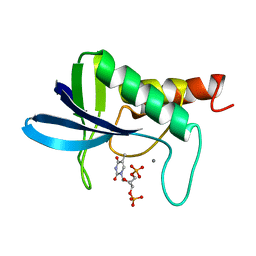 | | CRYSTAL STRUCTURES OF THE BINARY CA2+ AND PDTP COMPLEXES AND THE TERNARY COMPLEX OF THE ASP 21->GLU MUTANT OF STAPHYLOCOCCAL NUCLEASE. IMPLICATIONS FOR CATALYSIS AND LIGAND BINDING | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Libson, A, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-02-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding.
Biochemistry, 33, 1994
|
|
1ENE
 
 | |
1ENF
 
 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 1.69 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1ENH
 
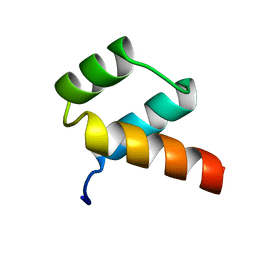 | | STRUCTURAL STUDIES OF THE ENGRAILED HOMEODOMAIN | | Descriptor: | ENGRAILED HOMEODOMAIN | | Authors: | Clarke, N.D, Kissinger, C.R, Desjarlais, J, Gilliland, G.L, Pabo, C.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the engrailed homeodomain.
Protein Sci., 3, 1994
|
|
1ENI
 
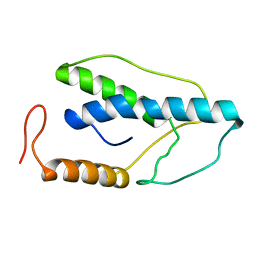 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENJ
 
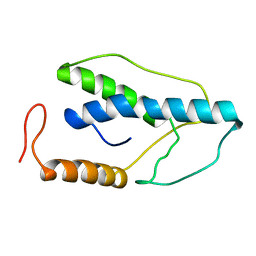 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
