2VU2
 
 | | Biosynthetic thiolase from Z. ramigera. Complex with S-pantetheine-11- pivalate. | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-({3-oxo-3-[(2-sulfanylethyl)amino]propyl}amino)butyl 2,2-dimethylpropanoate, ACETYL-COA ACETYLTRANSFERASE, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
5BZ4
 
 | | Crystal structure of a T1-like thiolase (CoA-complex) from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase, COENZYME A | | Authors: | Janardan, N, Harijan, R.K, Kiema, T.R, Wierenga, R.K, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1DLU
 
 | | UNLIGANDED BIOSYNTHETIC THIOLASE FROM ZOOGLOEA RAMIGERA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIOSYNTHETIC THIOLASE, SULFATE ION | | Authors: | Modis, Y, Wierenga, R.K. | | Deposit date: | 1999-12-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
J.Mol.Biol., 297, 2000
|
|
1DM3
 
 | |
4WYS
 
 | | Crystal structure of thiolase from Escherichia coli | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
5CBQ
 
 | |
4WYR
 
 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
1OU6
 
 | |
8BRJ
 
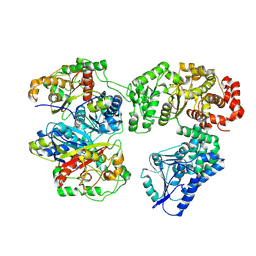 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
8BNU
 
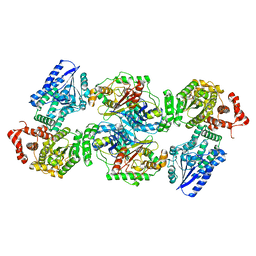 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
1DLV
 
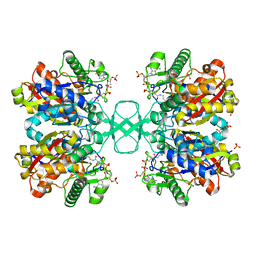 | |
8BNR
 
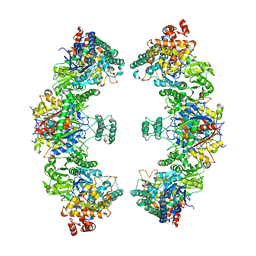 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) octameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
7CW5
 
 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
5BYV
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1PXT
 
 | |
7FEA
 
 | | PY14 in complex with Col-D | | Descriptor: | (6~{R},7~{R},9~{E})-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Ko, T.P, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
4XL4
 
 | | Crystal structure of thiolase from Clostridium acetobutylicum in complex with CoA | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL2
 
 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | Descriptor: | ACETATE ION, Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4O9A
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4O99
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4XL3
 
 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
6PCA
 
 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
6PCB
 
 | |
6PCC
 
 | |
6PCD
 
 | |
