2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
7N1N
 
 | | Prx in complex with ComR DNA-binding domain | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ComR, Prx | | Authors: | Rutbeek, N.R, Prehna, G. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of quorum sensing inhibition in Streptococcus by the phage protein paratox.
J.Biol.Chem., 297, 2021
|
|
7N10
 
 | |
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1SQ8
 
 | |
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
4OB4
 
 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
2XCJ
 
 | | Crystal structure of P2 C, the immunity repressor of temperate E. coli phage P2 | | Descriptor: | C PROTEIN, FORMIC ACID, GLYCEROL, ... | | Authors: | Massad, T, Skaar, K, Hogbom, M, Stenmark, P. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the P2 C-Repressor: A Binder of Non-Palindromic Direct DNA Repeats.
Nucleic Acids Res., 38, 2010
|
|
2XI8
 
 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
3KZ3
 
 | |
2XJ3
 
 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
3ZHI
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
3LFP
 
 | |
6H49
 
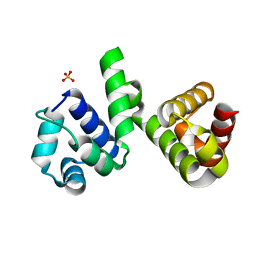 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
3LIS
 
 | |
6RNX
 
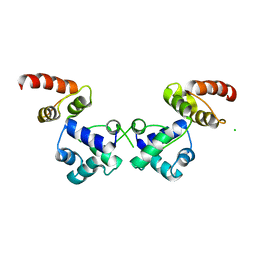 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
2XIU
 
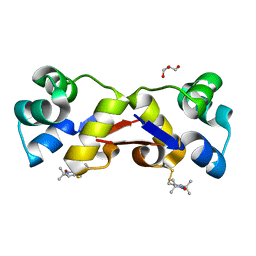 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
6B9S
 
 | |
4FN3
 
 | |
4FBI
 
 | |
4F8D
 
 | |
6RNZ
 
 | | Crystal structure of the N-terminal HTH DNA-binding domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
3ZHM
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 in complex with the OL2 operator half-site | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*AP*CP*TP*TP*GP*CP*AP*CP *TP*TP*GP*A)-3', 5'-D(*AP*GP*TP*TP*CP*AP*CP*GP*TP*TP*CP*AP*AP*GP *TP*GP*CP*A)-3', CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
6B9T
 
 | |
6RMQ
 
 | | Crystal structure of a selenomethionine-substituted A70M I84M mutant of the essential repressor DdrO from radiation resistant-Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
