4ISM
 
 | |
4ITW
 
 | |
2Q0Y
 
 | |
2Q14
 
 | |
4J0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(S)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
4IWR
 
 | | C.Esp1396I bound to a 25 base pair operator site | | Descriptor: | DNA (25-MER), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4J1P
 
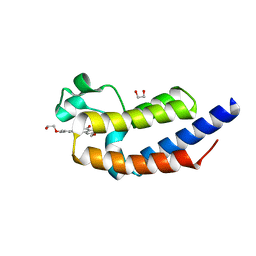 | | X-ray crystal structure of bromodomain 2 of human brd2 in complex with rvx208 to 1.08 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain containing 2, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-01 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
4IX8
 
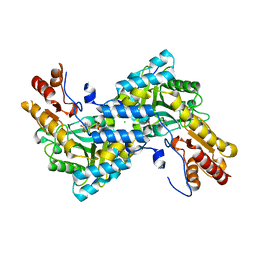 | |
4J27
 
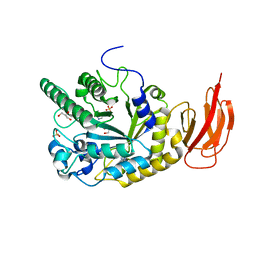 | |
3DB2
 
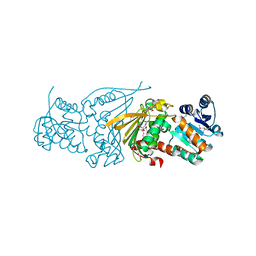 | |
4H5M
 
 | |
4H7A
 
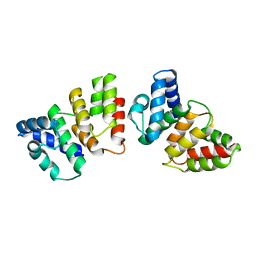 | | Crystal structure of CasB from Thermus thermophilus | | Descriptor: | CRISPR-associated protein Cse2 | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures.
Febs Lett., 586, 2012
|
|
4H8Y
 
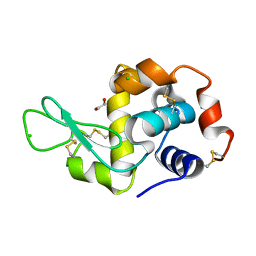 | | Radiation damage study of lysozyme- 0.14 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H93
 
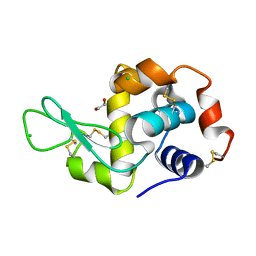 | | Radiation damage study of lysozyme - 0.49 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2003 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H9A
 
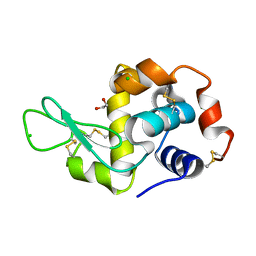 | | Radiation damage study of lysozyme - 0.63 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1997 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H9Y
 
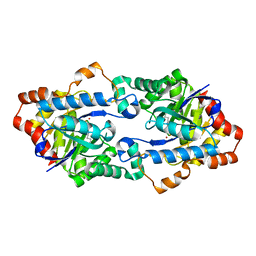 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H95
 
 | |
4H9E
 
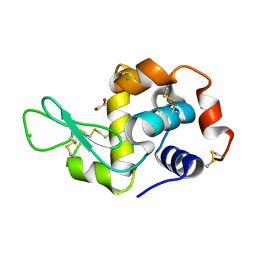 | | Radiation damage study of lysozyme - 0.84 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3DRG
 
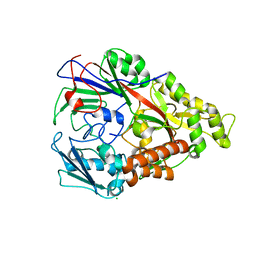 | | Lactococcal OppA complexed with bradykinin in the closed conformation | | Descriptor: | Bradykinin, CHLORIDE ION, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
4HE2
 
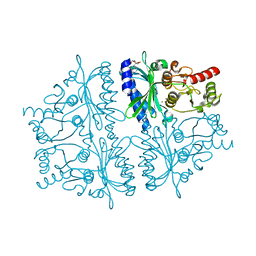 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3DOY
 
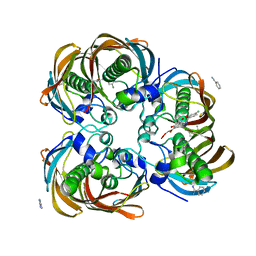 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3i | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-chloro-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
4H24
 
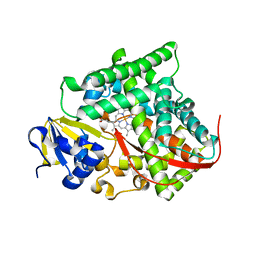 | | Cytochrome P450BM3-CIS cyclopropanation catalyst | | Descriptor: | Cytochrome P450-BM3 variant P450BM3-Cis, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Coelho, P.S, Wang, Z.J, Ener, M.E, Baril, S.A, Kannan, A, Arnold, F.H, Brustad, E.M. | | Deposit date: | 2012-09-11 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo.
Nat.Chem.Biol., 9, 2013
|
|
4H45
 
 | |
3DRK
 
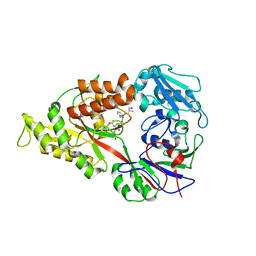 | | Crystal structure of Lactococcal OppA co-crystallized with Neuropeptide S in an open conformation | | Descriptor: | Neuropeptide S, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
4H2W
 
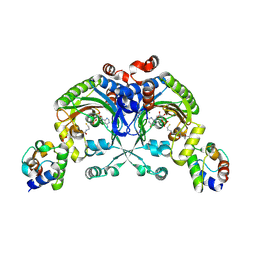 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
