1QBE
 
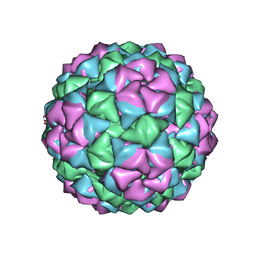 | | BACTERIOPHAGE Q BETA CAPSID | | Descriptor: | BACTERIOPHAGE Q BETA CAPSID | | Authors: | Liljas, L, Golmohammadi, R. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The crystal structure of bacteriophage Q beta at 3.5 A resolution.
Structure, 4, 1996
|
|
1MXA
 
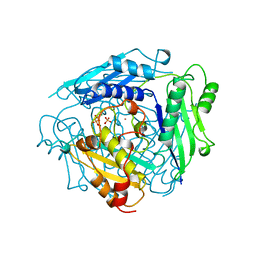 | | S-ADENOSYLMETHIONINE SYNTHETASE WITH PPI | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Markham, G.D. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of S-adenosylmethionine synthetase: crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution.
Biochemistry, 35, 1996
|
|
1WHI
 
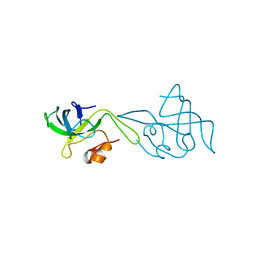 | | RIBOSOMAL PROTEIN L14 | | Descriptor: | RIBOSOMAL PROTEIN L14 | | Authors: | Davies, C, White, S.W, Ramakrishnan, V. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus.
Structure, 4, 1996
|
|
1FAJ
 
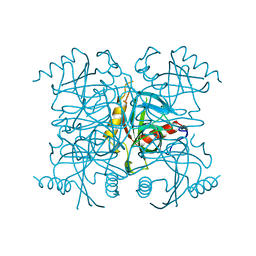 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1DKJ
 
 | | BOBWHITE QUAIL LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1MXC
 
 | | S-ADENOSYLMETHIONINE SYNTHETASE WITH 8-BR-ADP | | Descriptor: | 8-BROMOADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Markham, G.D. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of S-adenosylmethionine synthetase: crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution.
Biochemistry, 35, 1996
|
|
1XTC
 
 | | CHOLERA TOXIN | | Descriptor: | CHOLERA TOXIN | | Authors: | Zhang, R.-G, Westbrook, E. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional crystal structure of cholera toxin.
J.Mol.Biol., 251, 1995
|
|
2EIP
 
 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
243D
 
 | | STRUCTURE OF THE DNA OCTANUCLEOTIDE D(ACGTACGT)2 | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3') | | Authors: | Wilcock, D.J, Adams, A, Cardin, C.J, Wakelin, L.P.G. | | Deposit date: | 1996-01-10 | | Release date: | 1996-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DNA octanucleotide d(ACGTACGT)2.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MPO
 
 | | MALTOPORIN MALTOHEXAOSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1996-01-11 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway.
Structure, 4, 1996
|
|
1EXP
 
 | | BETA-1,4-GLYCANASE CEX-CD | | Descriptor: | BETA-1,4-D-GLYCANASE CEX-CD, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | White, A, Tull, D, Johns, K.L, Withers, S.G, Rose, D.R. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic observation of a covalent catalytic intermediate in a beta-glycosidase.
Nat.Struct.Biol., 3, 1996
|
|
1BOY
 
 | |
1ROO
 
 | | NMR SOLUTION STRUCTURE OF SHK TOXIN, NMR, 20 STRUCTURES | | Descriptor: | SHK TOXIN | | Authors: | Tudor, J.E, Pallaghy, P.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone.
Nat.Struct.Biol., 3, 1996
|
|
1RON
 
 | |
1MPM
 
 | | MALTOPORIN MALTOSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1996-01-11 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway.
Structure, 4, 1996
|
|
1MPN
 
 | | MALTOPORIN MALTOTRIOSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1996-01-11 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway.
Structure, 4, 1996
|
|
1KUL
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, 5 STRUCTURES | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KUM
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1CZJ
 
 | | CYTOCHROME C OF CLASS III (AMBLER) 26 KD | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Czjzek, M, Haser, R. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a dimeric octaheme cytochrome c3 (M(r) 26,000) from Desulfovibrio desulfuricans Norway.
Structure, 4, 1996
|
|
245D
 
 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1996-01-12 | | Release date: | 1996-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MIR
 
 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
1WAN
 
 | | DNA DTA TRIPLEX, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*AP*GP*AP*AP*CP*CP*CP*CP*TP*TP*CP*TP*AP*TP*CP*TP*TP*AP*TP*AP*TP*CP*TP*(D3)P*TP*CP*TP*T)-3') | | Authors: | Wang, E, Koshlap, K.M, Gillespie, P, Dervan, P.B, Feigon, J. | | Deposit date: | 1996-01-14 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pyrimidine-purine-pyrimidine triplex containing the sequence-specific intercalating non-natural base D3.
J.Mol.Biol., 257, 1996
|
|
1OBP
 
 | | ODORANT-BINDING PROTEIN FROM BOVINE NASAL MUCOSA | | Descriptor: | ODORANT-BINDING PROTEIN, UNKNOWN ATOM OR ION | | Authors: | Tegoni, M, Cambillau, C. | | Deposit date: | 1996-01-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain swapping creates a third putative combining site in bovine odorant binding protein dimer
Nat.Struct.Biol., 3, 1996
|
|
1ZAP
 
 | | SECRETED ASPARTIC PROTEASE FROM C. ALBICANS | | Descriptor: | N-ethyl-N-[(4-methylpiperazin-1-yl)carbonyl]-D-phenylalanyl-N-[(1S,2S,4R)-4-(butylcarbamoyl)-1-(cyclohexylmethyl)-2-hydroxy-5-methylhexyl]-L-norleucinamide, SECRETED ASPARTIC PROTEINASE, ZINC ION | | Authors: | Abad-Zapatero, C, Muchmore, S.W. | | Deposit date: | 1996-01-16 | | Release date: | 1997-04-21 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents.
Protein Sci., 5, 1996
|
|
1YJB
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 35% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
