1YTT
 
 | | YB SUBSTITUTED SUBTILISIN FRAGMENT OF MANNOSE BINDING PROTEIN-A (SUB-MBP-A), MAD STRUCTURE AT 110K | | Descriptor: | MANNOSE-BINDING PROTEIN A, YTTERBIUM (III) ION | | Authors: | Burling, F.T, Weis, W.I, Flaherty, K.M, Brunger, A.T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-06-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of protein solvation and discrete disorder with experimental crystallographic phases.
Science, 271, 1996
|
|
1LLP
 
 | | LIGNIN PEROXIDASE (ISOZYME H2) PI 4.15 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Choinowski, T.H, Piontek, K, Glumoff, T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of lignin peroxidase at 1.70 A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle.
J.Mol.Biol., 286, 1999
|
|
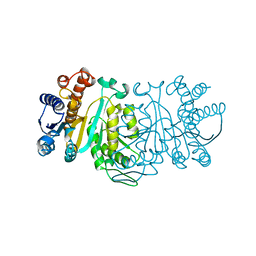
1XAB
 
 | |
1XAA
 
 | |
1XAC
 
 | |
1XAD
 
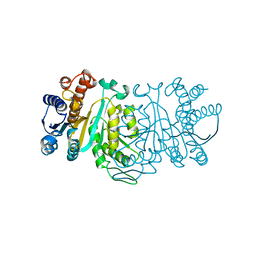 | |
1RTF
 
 | |
1LKK
 
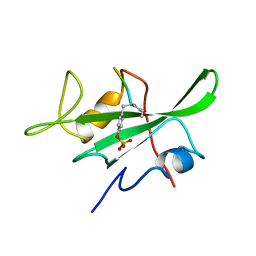 | |
1LKL
 
 | |
1QUJ
 
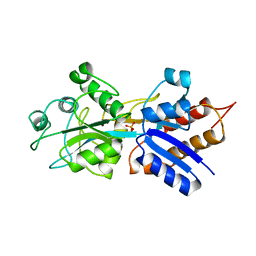
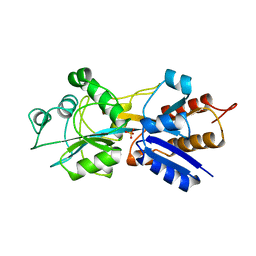 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUK
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY ASN COMPLEX WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUL
 
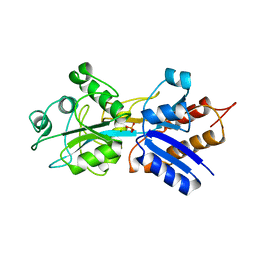 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY THR COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1BTM
 
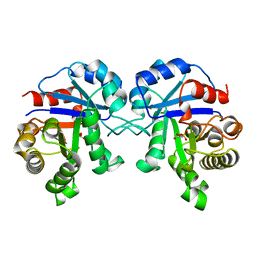 | | TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Delboni, L.F, Mande, S.C, Hol, W.G.J. | | Deposit date: | 1995-11-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci., 4, 1995
|
|
1CER
 
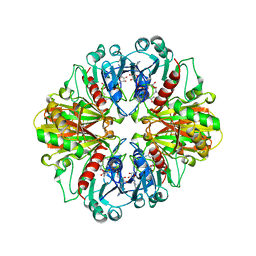 | |
1QUI
 
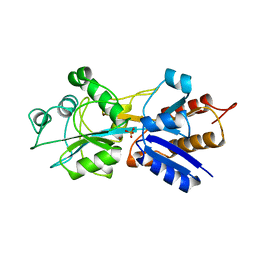 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH BROMINE AND PHOSPHATE | | Descriptor: | BROMIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
2BTA
 
 | |
2BTB
 
 | |
1VTP
 
 | |
1BMN
 
 | |
1BMM
 
 | |
1DRN
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GGAGA)R(UGAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*GP*AP*GP*A)-R(P*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-11-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
1PEA
 
 | |
1CUW
 
 | | CUTINASE, G82A, A85F, V184I, A185L, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUE
 
 | | CUTINASE, Q121L MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUC
 
 | |
