1XIU
 
 | | Crystal structure of the agonist-bound ligand-binding domain of Biomphalaria glabrata RXR | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 1, RXR-like protein | | Authors: | De Groot, A, De Rosny, E, Juillan-Binard, C, Ferrer, J.-L, Laudet, V, Pebay-Peroula, E, Fontecilla-Camps, J.-C, Borel, F. | | Deposit date: | 2004-09-22 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Novel Tetrameric Complex of Agonist-bound Ligand-binding Domain of Biomphalaria glabrata Retinoid X Receptor.
J.Mol.Biol., 354, 2005
|
|
6B4R
 
 | | The crystal structure of the aldehyde dehydrogenase KauB from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Cardona-Cardona, Y, Carrillo-Campos, J, Munoz-Clares, R.A. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Aldehyde specificity of the aldehyde dehydrogenase KauB from Pseudomonas aeruginosa: Critical amino acid residues revealed by its crystal structure
To Be Published
|
|
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
2B3Y
 
 | | Structure of a monoclinic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
3TAL
 
 | | Crystal structure of NurA with manganese | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL, MANGANESE (II) ION | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3WRI
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of P450cam intermediate
To be Published
|
|
3WRM
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
2HMQ
 
 | |
2HMZ
 
 | |
3EVP
 
 | | crystal structure of circular-permutated EGFP | | Descriptor: | Green fluorescent protein,Green fluorescent protein | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3WRJ
 
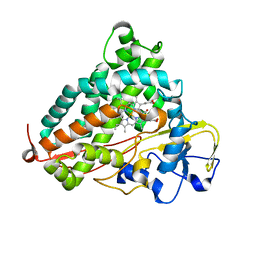 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of P450cam intermedite
to be published
|
|
3WRL
 
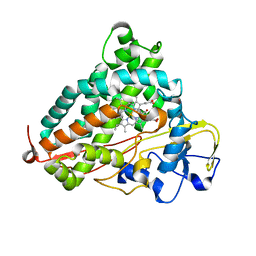 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
4EB5
 
 | | A. fulgidus IscS-IscU complex structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Marinoni, E.N, de Oliveira, J.S, Nicolet, Y, Raulfs, E.C, Amara, P, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-03-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7NWR
 
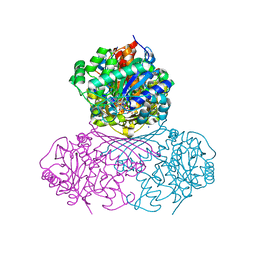 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
1D63
 
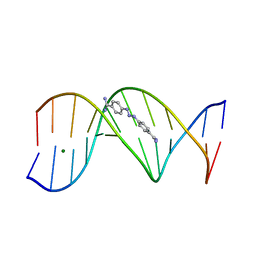 | | CRYSTAL STRUCTURE OF A BERENIL-D(CGCAAATTTGCG) COMPLEX; AN EXAMPLE OF DRUG-DNA RECOGNITION BASED ON SEQUENCE-DEPENDENT STRUCTURAL FEATURES | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Brown, D.G, Sanderson, M.R, Garman, E, Neidle, S. | | Deposit date: | 1992-03-02 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features.
J.Mol.Biol., 226, 1992
|
|
1CFG
 
 | |
7NZY
 
 | | Crystal structure of human Casein Kinase I delta in complex with CGS-15943 | | Descriptor: | 9-chloranyl-2-(furan-2-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5-amine, Casein kinase I isoform delta, MALONATE ION, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2021-03-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phenotypic Discovery of Triazolo[1,5- c ]quinazolines as a First-In-Class Bone Morphogenetic Protein Amplifier Chemotype.
J.Med.Chem., 65, 2022
|
|
5CP4
 
 | | CRYOGENIC STRUCTURE OF P450CAM | | Descriptor: | CAMPHOR, CYTOCHROME P450CAM, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-05-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect.
Biochemistry, 37, 1998
|
|
8S24
 
 | |
4F3H
 
 | | The structural of FimXEAL-c-di-GMP from Xanthomonas campestris | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein | | Authors: | Chin, K.-H, Liao, Y.-T, Chou, S.-H. | | Deposit date: | 2012-05-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1X3M
 
 | |
4F48
 
 | | The X-ray structural of FimXEAL-c-di-GMP-PilZ complexes from Xanthomonas campestris | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein, Type IV fimbriae assembly protein | | Authors: | Chin, K.-H, Liao, Y.-T, Chou, S.-H. | | Deposit date: | 2012-05-10 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7Q71
 
 | | The crystallographic structure of the Ligand Binding domain of the NR7 nuclear receptor from the amphioxus Branchiostoma lanceolatum | | Descriptor: | CHLORIDE ION, Nuclear hormone receptor 7, PHOSPHATE ION | | Authors: | Billas, I.M.L, McEwen, A.G, Hazemann, I, Moras, D, Laudet, V. | | Deposit date: | 2021-11-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel nuclear receptor subfamily enlightens the origin of heterodimerization.
Bmc Biol., 20, 2022
|
|
3T9V
 
 | |
