9CIL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T41V/S59A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T41V/S59A at cryogenic temperature
To Be Published
|
|
9CIK
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, SIegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature
To Be Published
|
|
9CIJ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, L.J, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature
To Be Published
|
|
9CII
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, Robinson, A.C, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36R at cryogenic temperature
To Be Published
|
|
9CIA
 
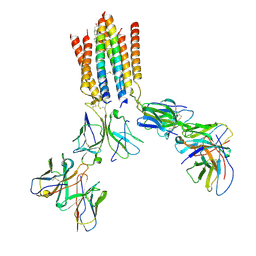 | | T cell receptor complex | | Descriptor: | CHOLESTEROL, T cell receptor delta constant, T cell receptor gamma constant 1, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
9CI8
 
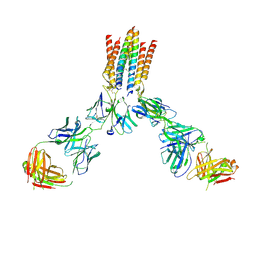 | | T cell receptor complex | | Descriptor: | T cell receptor delta constant, T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-02 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
9CGT
 
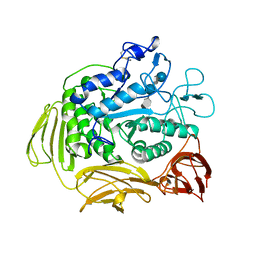 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOPENTAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-1-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
9CGM
 
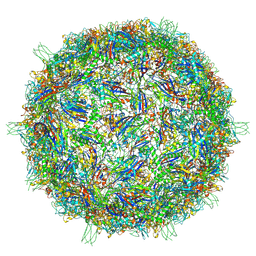 | | The Structure of Spiroplasma Virus 4 | | Descriptor: | Capsid protein VP1, DNA binding protein ORF8 | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The Structure of Spiroplasma Virus 4 : Exploring the Capsid Diversity of the Microviridae.
Viruses, 16, 2024
|
|
9CEQ
 
 | |
9CE9
 
 | |
9CC3
 
 | | Human Mitochondrial LONP1 Stall State + casein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endogenous Co-purified substrate modeled as unknown residues, Lon protease homolog, ... | | Authors: | Mindrebo, J.T, Lander, G.C. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural and mechanistic studies on human LONP1 redefine the hand-over-hand translocation mechanism.
Biorxiv, 2024
|
|
9CC0
 
 | |
9CBT
 
 | | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-6-[[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-6-oxidanyl-2-tridecyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]amino]-1-oxidanylidene-1-[2-(triethyl-$l^{4}-azanyl)ethylamino]hexan-2-yl]carbamate, 2-{[(2S)-6-[(Z)-(1-{[(2R,3R,4R,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-4-hydroxy-2-sulfanyloxolan-3-yl]oxy}tetradecylidene)amino]-2-{[(benzyloxy)carbonyl]amino}hexanoyl]amino}-N,N,N-triethylethan-1-aminium (non-preferred name), NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Fenwick, M.K, Young, H.J, Lin, H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10
To Be Published
|
|
9CBE
 
 | |
9CA2
 
 | |
9CA1
 
 | | Human TOP3B-TDRD3 core complex in DNA religation state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
9CA0
 
 | | Human TOP3B-TDRD3 core complex in DNA pre-cleavage state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
9C9Y
 
 | | Human TOP3B-TDRD3 core complex in DNA pre-cleavage state | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*T)-3'), DNA topoisomerase 3-beta-1, MANGANESE (II) ION, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
9C9M
 
 | | HIV-1 intasome core bound with DTG | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, Integrase, MAGNESIUM ION, ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.01 Å) | | Cite: | HIV-1 Intasomes Assembled with Excess Integrase C-Terminal Domain Protein Facilitate Structural Studies by Cryo-EM and Reveal the Role of the Integrase C-Terminal Tail in HIV-1 Integration.
Viruses, 16, 2024
|
|
9C8S
 
 | |
9C8R
 
 | |
9C8J
 
 | |
9C89
 
 | |
9C84
 
 | |
9C83
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | AmpC Beta-lactamase, N-[(3M)-3-(5-chloro-1,2,3-thiadiazol-4-yl)phenyl]-5-methyl-3-oxo-2,3-dihydro-1,2-oxazole-4-sulfonamide | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
