2MVY
 
 | |
2M24
 
 | |
4E2V
 
 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with preQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Schmidt, I, Reuter, K, Klebe, G, Heine, A, Tidten, N. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
5YN4
 
 | |
5TKO
 
 | | RNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-10-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | RNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
2MER
 
 | |
2MEQ
 
 | |
2M39
 
 | |
2N1Q
 
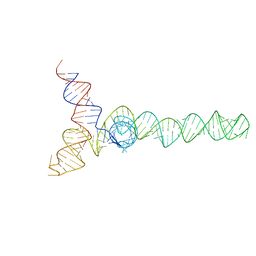 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
1RB8
 
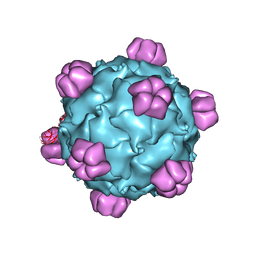 | | The phiX174 DNA binding protein J in two different capsid environments. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Capsid protein, DNA (5'-D(P*CP*AP*AP*A)-3'), ... | | Authors: | Bernal, R.A, Hafenstein, S, Esmeralda, R, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2003-11-03 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The phiX174 Protein J Mediates DNA Packaging and Viral Attachment to Host Cells.
J.Mol.Biol., 337, 2004
|
|
5XWG
 
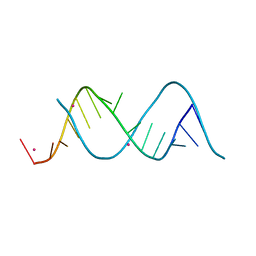 | |
5N5C
 
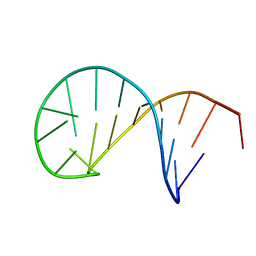 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
8SLD
 
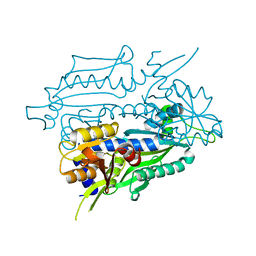 | |
5V0K
 
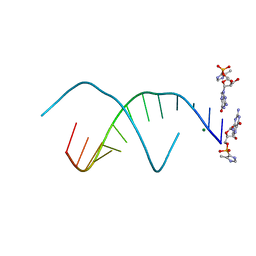 | | RNA duplex with 2-MeImpG analogue bound-3 binding sites | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*C)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA duplex with 2-MeImpG analogue bound-3 binding sites
To Be Published
|
|
2RVO
 
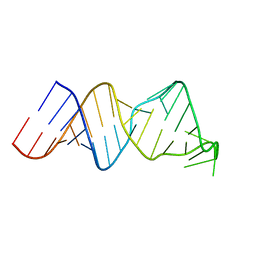 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | Descriptor: | RNA (34-MER) | | Authors: | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
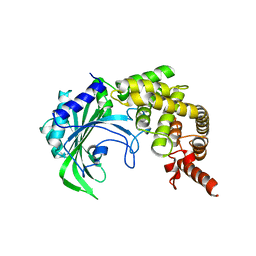
3HTZ
 
 | |
2M12
 
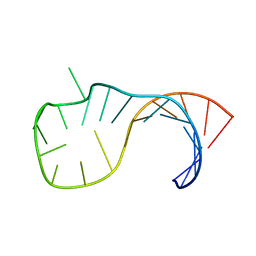 | |
3NFF
 
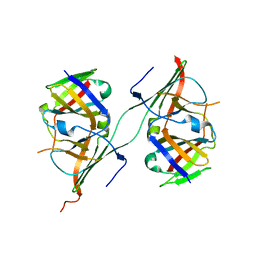 | | Crystal structure of extended Dimerization module of RNA polymerase I subcomplex A49/A34.5 | | Descriptor: | RNA polymerase I subunit A34.5, RNA polymerase I subunit A49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
3BLD
 
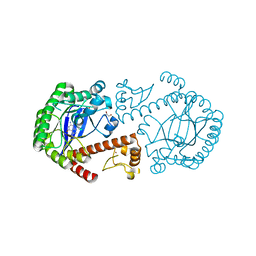 | | tRNA guanine transglycosylase V233G mutant preQ1 complex structure | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
3BL3
 
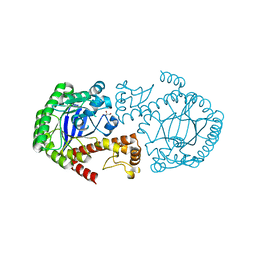 | | tRNA guanine transglycosylase V233G mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-10 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
409D
 
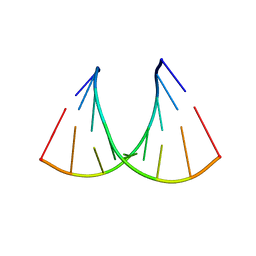 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | Descriptor: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | Authors: | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
2L8C
 
 | |
2JXQ
 
 | | NMR structure of RNA duplex | | Descriptor: | RNA (5'-R(*CP*GP*CP*UP*CP*UP*CP*UP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Popenda, L, Adamiak, R.W, Gdaniec, Z. | | Deposit date: | 2007-11-28 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Bulged adenosine influence on the RNA duplex conformation in solution.
Biochemistry, 47, 2008
|
|
1JKE
 
 | | D-Tyr tRNATyr deacylase from Escherichia coli | | Descriptor: | D-Tyr-tRNATyr deacylase, ZINC ION | | Authors: | Ferri-Fioni, M.L, Schmitt, E, Soutourina, J, Plateau, P, Mechulam, Y, Blanquet, S. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of crystalline D-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases.
J.Biol.Chem., 276, 2001
|
|
1UUU
 
 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
