3PCA
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCI
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-IODO-4-HYDROXYBENZOATE | | Descriptor: | 3-IODO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCN
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3,4-DIHYDROXYPHENYLACETATE | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and resonance Raman studies of protocatechuate 3,4-dioxygenase complexed with 3,4-dihydroxyphenylacetate.
Biochemistry, 36, 1997
|
|
3PCK
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCM
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCH
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-CHLORO-4-HYDROXYBENZOATE | | Descriptor: | 3-CHLORO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCJ
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCL
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3U8M
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3920 (1-(6-bromopyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
2WAR
 
 | | Hen Egg White Lysozyme E35Q chitopentaose complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME C | | Authors: | Davies, G.J, Withers, S.G, Vocadlo, D.J. | | Deposit date: | 2009-02-13 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Chitopentaose Complex of a Mutant Hen Egg-White Lysozyme Displays No Distortion of the -1 Sugar Away from a 4C1 Chair Conformation
Aust.J.Chem., 62, 2009
|
|
2WBK
 
 | | Structure of the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-mannopyranoside, BETA-MANNOSIDASE, ... | | Authors: | Offen, W.A, Zechel, D.L, Withers, S.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Michaelis Complex of Beta-Mannosidase, Man2A, Provides Insight Into the Conformational Itinerary of Mannoside Hydrolysis.
Cell(Cambridge,Mass.), 18, 2009
|
|
3U7B
 
 | | A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of a GH10 xylanase from Fusarium oxysporum reveals the presence of an extended loop on top of the catalytic cleft.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TZH
 
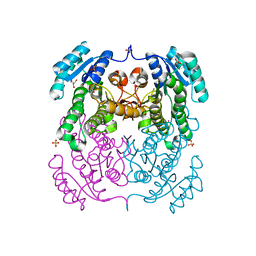 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(F187A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, SULFATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Osinski, T, Shumilin, I, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
2VXI
 
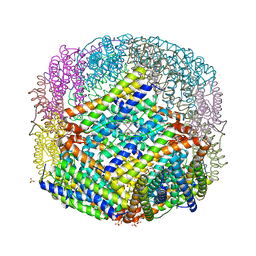 | | The binding of heme and zinc in Escherichia coli Bacterioferritin | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Willies, S.C, Isupov, M.N, Garman, E.F, Littlechild, J.A. | | Deposit date: | 2008-07-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Binding of Haem and Zinc in the 1.9 A X-Ray Structure of Escherichia Coli Bacterioferritin.
J.Biol.Inorg.Chem., 14, 2009
|
|
3IE2
 
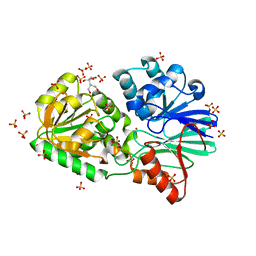 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IDZ
 
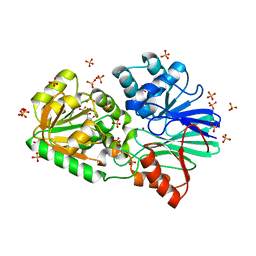 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3C3P
 
 | |
3C5Y
 
 | |
3U79
 
 | | AL-103 Y32F Y96F | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Amyloidogenic immunoglobulin light chain protein AL-103 Y32F Y96F, ... | | Authors: | DiCostanzo, A.C, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Tyrosine Residues mediate crucial interactions in amyloid formation for immunoglobulin light chains
To be Published
|
|
3QKI
 
 | | Crystal structure of Glucose-6-Phosphate Isomerase (PF14_0341) from Plasmodium falciparum 3D7 | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Gileadi, T, Wernimont, A.K, Hutchinson, A, Weadge, J, Cossar, D, Lew, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Glucose-6-Phosphate Isomerase (PF14_0341) from Plasmodium falciparum 3D7
TO BE PUBLISHED
|
|
3U9F
 
 | |
3UBJ
 
 | | Influenza hemagglutinin from the 2009 pandemic in complex with ligand LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Characterization of the Hemagglutinin Receptor Specificity from the 2009 H1N1 Influenza Pandemic.
J.Virol., 86, 2012
|
|
3IOS
 
 | | Structure of MTB dsbF in its mixed oxidized and reduced forms | | Descriptor: | Disulfide bond forming protein (DsbF) | | Authors: | Chim, N, Goulding, C.W. | | Deposit date: | 2009-08-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An extracellular disulfide bond forming protein (DsbF) from Mycobacterium tuberculosis: structural, biochemical, and gene expression analysis.
J.Mol.Biol., 396, 2010
|
|
3ISE
 
 | | Structure of mineralized Bfrb (double soak) from Pseudomonas aeruginosa to 2.8A Resolution | | Descriptor: | Bacterioferritin, FE (III) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
2R9S
 
 | | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 10, N-(tert-butyl)-4-[5-(pyridin-2-ylamino)quinolin-3-yl]benzenesulfonamide, ... | | Authors: | Habel, J. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
