3QQ5
 
 | | Crystal structure of the [FeFe]-hydrogenase maturation protein HydF | | Descriptor: | Small GTP-binding protein | | Authors: | Cendron, L, Berto, P, D'Adamo, S, Vallese, F, Govoni, C, Posewitz, M.C, Giacometti, G.M, Costantini, P, Zanotti, G. | | Deposit date: | 2011-02-15 | | Release date: | 2011-11-16 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of HydF Scaffold Protein Provides Insights into [FeFe]-Hydrogenase Maturation.
J.Biol.Chem., 286, 2011
|
|
3QTZ
 
 | | CDK2 in complex with inhibitor RC-2-36 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(3-fluorobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3QN7
 
 | | Potent and selective bicyclic peptide inhibitor (UK18) of human urokinase-type plasminogen activator(uPA) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, Bicyclic peptide inhibitor, Urokinase-type plasminogen activator | | Authors: | Angelini, A, Cendron, L, Touati, J, Winter, G, Zanotti, G, Heinis, C. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bicyclic peptide inhibitor reveals large contact interface with a protease target
Acs Chem.Biol., 7, 2012
|
|
3QTU
 
 | | CDK2 in complex with inhibitor RC-2-132 | | Descriptor: | 4-{[4-amino-5-(4-sulfamoylbenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3QTQ
 
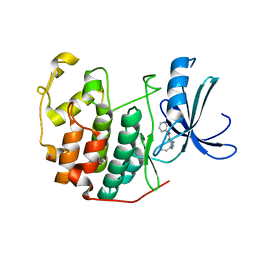 | | CDK2 in complex with inhibitor RC-1-137 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](pyridin-3-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3QTW
 
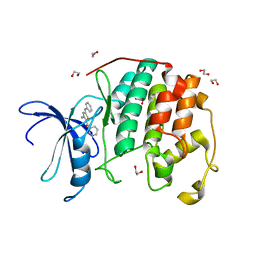 | | CDK2 in complex with inhibitor RC-2-13 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(phenylamino)-1,3-thiazol-5-yl](pyridin-3-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RKB
 
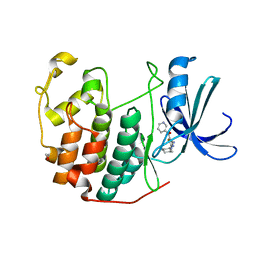 | | CDK2 in complex with inhibitor RC-2-73 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(cyclohexylamino)-1,3-thiazol-5-yl](pyridin-3-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3S0O
 
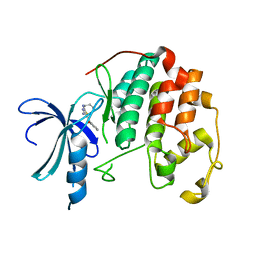 | | CDK2 in complex with inhibitor RC-1-138 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](pyridin-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-05-13 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3SQQ
 
 | | CDK2 in complex with inhibitor RC-3-96 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-methylbenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-07-06 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RAL
 
 | | CDK2 in complex with inhibitor RC-2-34 | | Descriptor: | 4-{[4-amino-5-(3-methoxybenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RAH
 
 | | CDK2 in complex with inhibitor RC-2-22 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(cyclohexylamino)-1,3-thiazol-5-yl](naphthalen-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3TKH
 
 | | Crystal structure of Chk1 in complex with inhibitor S01 | | Descriptor: | 1-(morpholin-4-yl)-2-[4-(2-{[5-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}pyridin-4-yl)piperazin-1-yl]ethanone, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TLM
 
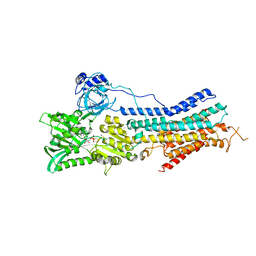 | | Crystal Structure of Endoplasmic Reticulum Ca2+-ATPase (SERCA) From Bovine Muscle | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sacchetto, R, Bertipaglia, I, Giannetti, S, Cendron, L, Mascarello, F, Damiani, E, Carafoli, E, Zanotti, G. | | Deposit date: | 2011-08-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of sarcoplasmic reticulum Ca(2+)-ATPase (SERCA) from bovine muscle.
J.Struct.Biol., 178, 2012
|
|
3RJC
 
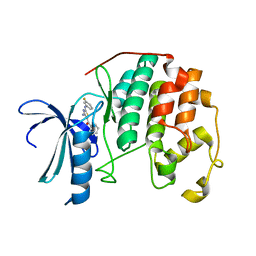 | | CDK2 in complex with inhibitor L4-12 | | Descriptor: | Cyclin-dependent kinase 2, {4-amino-2-[(3-fluorophenyl)amino]-1,3-thiazol-5-yl}(phenyl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3TKI
 
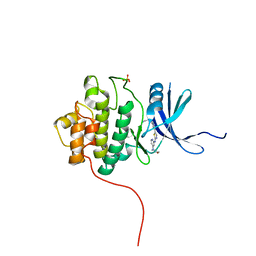 | | Crystal structure of Chk1 in complex with inhibitor S25 | | Descriptor: | N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TKD
 
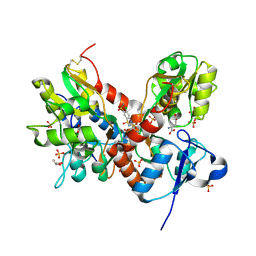 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and cyclothiazide at 1.45 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
3R8U
 
 | | CDK2 in complex with inhibitor RC-1-132 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](naphthalen-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3R9O
 
 | | CDK2 in complex with inhibitor RC-2-143 | | Descriptor: | 4-amino-N-(3,5-difluorophenyl)-2-[(4-sulfamoylphenyl)amino]-1,3-thiazole-5-carboxamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
5CCT
 
 | | Staphylococcus bacteriophage 80alpha dUTPase G164S mutant with dUpNHpp. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Maiques, E, Quiles-Puchalt, N, Donderis, J, Ciges, J.R, Alite, C, Bowring, J, Penades, J.R, Marina, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party.
Nucleic Acids Res., 44, 2016
|
|
3AOV
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein PH0207 | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-10-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of kynurenine aminotransferase from hyperthermophilic archaeon: enzymatic activity for conversion to kynurenic acid is allosterically regulated by alpha-ketoglutaric acid cooperatively with kynurenine
To be Published
|
|
5CCO
 
 | | Staphylococcus bacteriophage 80alpha dUTPase with dUMP. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Maiques, E, Quiles-Puchalt, N, Donderis, J, Ciges, J.R, Alite, C, Bowring, J, Penades, J.R, Marina, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party.
Nucleic Acids Res., 44, 2016
|
|
6FLS
 
 | |
2XWH
 
 | | HCV-J6 NS5B polymerase structure at 1.8 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, POLYETHYLENE GLYCOL (N=34), RNA DEPENDENT RNA POLYMERASE | | Authors: | Scrima, N, Bressanelli, S. | | Deposit date: | 2010-11-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
2XYM
 
 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
2VJ2
 
 | | Human Jagged-1, domains DSL and EGFs1-3 | | Descriptor: | D-MALATE, JAGGED-1 | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Handford, P.A, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
