8QI3
 
 | | 390A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9EOO
 
 | |
9EOM
 
 | |
9EON
 
 | |
9EOP
 
 | |
4IL5
 
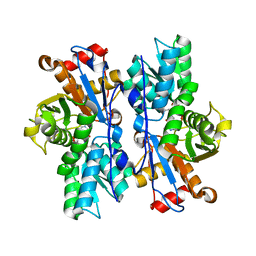 | |
4JBN
 
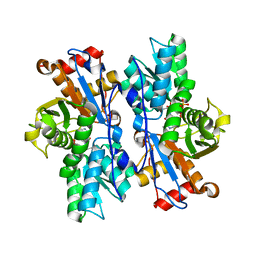 | |
5LXH
 
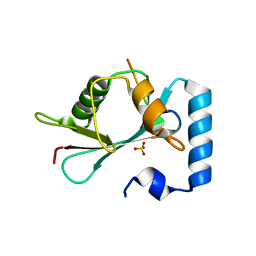 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | Cysteine protease ATG4B, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
4JBL
 
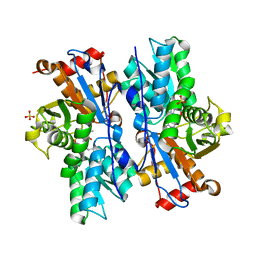 | |
9ILX
 
 | | Thanatin VF16QK in complex with LPS | | Descriptor: | VAL-PRO-ILE-ILE-TYR-CYS-ASN-ARG-ARG-THR-DLY-LYS-CYS-LYS-ARG-PHE | | Authors: | Swaleeha, A, Bhattacharyya, S. | | Deposit date: | 2024-07-01 | | Release date: | 2025-05-07 | | Method: | SOLUTION NMR | | Cite: | Single Disulfide Bond in Host Defense Thanatin Analog Peptides: Antimicrobial Activity, Atomic-Resolution Structures and Target Interactions.
Int J Mol Sci, 26, 2024
|
|
9IKO
 
 | | VF16QK in free solution | | Descriptor: | VAL-PRO-ILE-ILE-TYR-CYS-ASN-ARG-ARG-THR-DLY-LYS-CYS-LYS-ARG-PHE | | Authors: | Swaleeha, A, Bhattacharyya, S. | | Deposit date: | 2024-06-28 | | Release date: | 2025-05-07 | | Method: | SOLUTION NMR | | Cite: | Single Disulfide Bond in Host Defense Thanatin Analog Peptides: Antimicrobial Activity, Atomic-Resolution Structures and Target Interactions.
Int J Mol Sci, 26, 2024
|
|
9MTR
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site GGS-mutant Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTQ
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site GGD-mutant Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTS
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-unmodified Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTP
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site fMEAAAKC-peptidyl-tRNAcys at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
9MTT
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site deacylated-tRNAcys at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
1KO2
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with an oxidized Cys (cysteinesulfonic) | | Descriptor: | ACETATE ION, VIM-2 metallo-beta-lactamase, ZINC ION | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
9NO7
 
 | | Cryo-EM structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site 2'-deoxy-A76-fMEAAAKC-peptidyl-tRNAcys at 2.13A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-03-08 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
3JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
6EXO
 
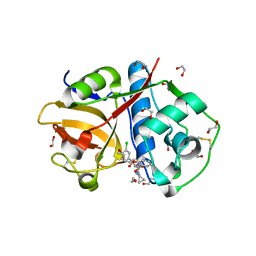 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6Z7R
 
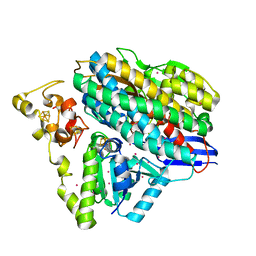 | | Structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris hildenborough pressurized with Krypton gas - structure wtKr1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Zacarias, S, Temporao, A, Carpentier, P, van der Linden, P, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
J.Biol.Inorg.Chem., 25, 2020
|
|
3DSW
 
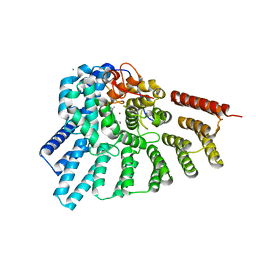 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys(GG)-Ser-Cys derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSX
 
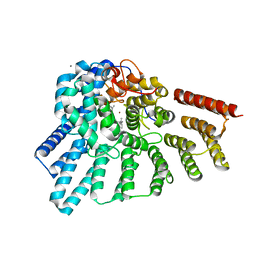 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with di-prenylated peptide Ser-Cys(GG)-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSV
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
1LLM
 
 | |
