5HBS
 
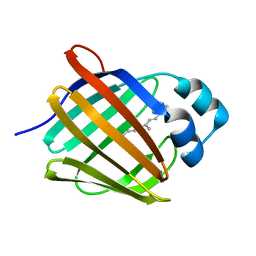 | | Crystal structure of human cellular retinol binding protein 1 in complex with all-trans-retinol at 0.89 angstrom. | | Descriptor: | RETINOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2016-01-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
1AA5
 
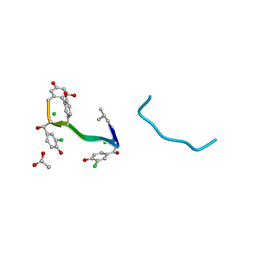 | | VANCOMYCIN | | Descriptor: | ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Bevivino, A.E, Korty, B.D, Axelsen, P.H. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Simultaneous Recognition of a Carboxylate-Containing Ligand and an Intramolecular Surrogate Ligand in the Crystal Structure of an Asymmetric Vancomycin Dimer.
J.Am.Chem.Soc., 119, 1997
|
|
1OB7
 
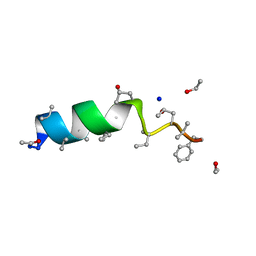 | | Cephaibol C | | Descriptor: | CEPHAIBOL C, ETHANOL, SODIUM ION | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
1ETN
 
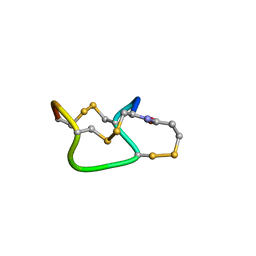 | |
8AVU
 
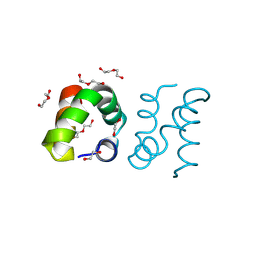 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the dimeric state | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
1JXW
 
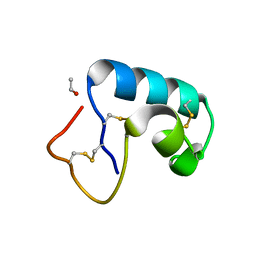 | | CRAMBIN MIXED SEQUENCE FORM AT 180 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7TVL
 
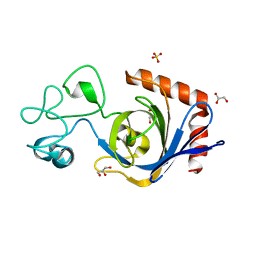 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
1I1W
 
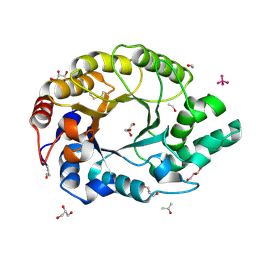 | | 0.89A Ultra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus | | Descriptor: | ACETONE, ENDO-1,4-BETA-XYLANASE, ETHANOL, ... | | Authors: | Natesh, R, Ramakumar, S, Viswamitra, M.A. | | Deposit date: | 2001-02-04 | | Release date: | 2003-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Thermostable xylanase from Thermoascus aurantiacus at ultrahigh resolution (0.89 A) at 100 K and atomic resolution (1.11 A) at 293 K refined anisotropically to small-molecule accuracy.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1JXT
 
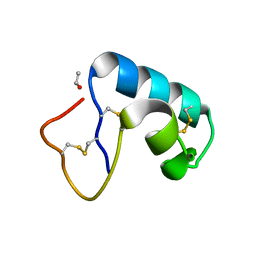 | | CRAMBIN MIXED SEQUENCE FORM AT 160 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXY
 
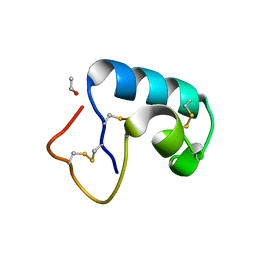 | | CRAMBIN MIXED SEQUENCE FORM AT 220 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXX
 
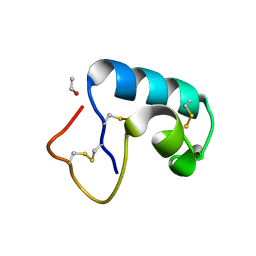 | | CRAMBIN MIXED SEQUENCE FORM AT 200 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1YWA
 
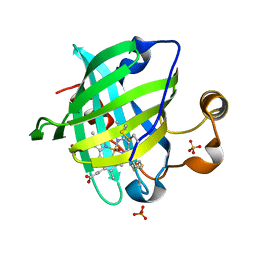 | | 0.9 A Structure of NP4 from Rhodnius Prolixus complexed with CO at pH 5.6 | | Descriptor: | CARBON MONOXIDE, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
2QDV
 
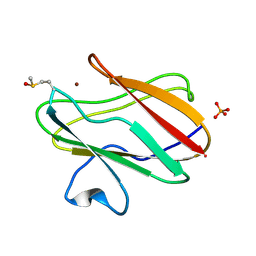 | | Structure of the Cu(II) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Wang, Y, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
1HHU
 
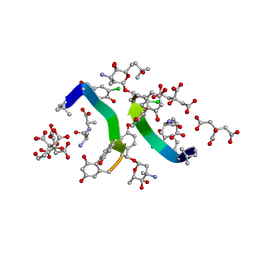 | | Balhimycin in complex with D-Ala-D-Ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, BALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertessy, L. | | Deposit date: | 2000-12-28 | | Release date: | 2003-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
4WEE
 
 | |
2YL7
 
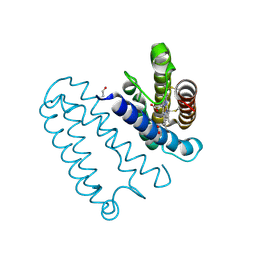 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: AS ISOLATED L16G VARIANT AT 0.9 A RESOLUTION - RESTRAINT REFINEMENT | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XU3
 
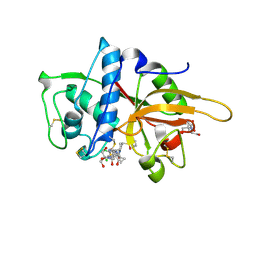 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-1-[1-(5-chlorothiophen-2-yl)cyclopropyl]carbonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATHEPSIN L1, ... | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
1M24
 
 | |
5R32
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 26, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
8SH8
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain Asn40Asp mutant in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Papain-like protease nsp3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2023-04-13 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 NSP3 macrodomain Asn40Asp mutant in complex with ADP-ribose (P43 crystal form)
To Be Published
|
|
8SH6
 
 | |
5X9L
 
 | | Recombinant thaumatin I at 0.9 Angstrom | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
6KLZ
 
 | | Human Carbonic Anhydrase II V143I variant 00 atm CO2 | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
4TUT
 
 | | Structure of a Prion peptide | | Descriptor: | Prion peptide: GLY-GLY-TYR-MET-LEU-GLY | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
6KKZ
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 8.5 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
