1CKD
 
 | | T43V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKE
 
 | | CMP KINASE FROM ESCHERICHIA COLI FREE ENZYME STRUCTURE | | Descriptor: | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Briozzo, P, Golinelli-Pimpaneau, B. | | Deposit date: | 1998-09-24 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
1CKF
 
 | | T52A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKG
 
 | | T52V MUTANT HUMAN LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKH
 
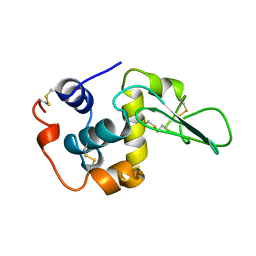 | | T70V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKI
 
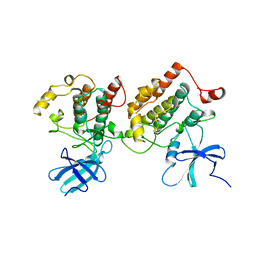 | |
1CKJ
 
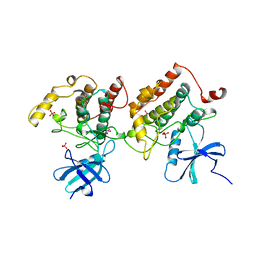 | |
1CKK
 
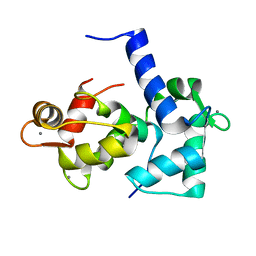 | | CALMODULIN/RAT CA2+/CALMODULIN DEPENDENT PROTEIN KINASE FRAGMENT | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase kinase 1, Calmodulin-1 | | Authors: | Osawa, M, Tokumitsu, H, Swindells, M.B, Kurihara, H, Orita, M, Shibanuma, T, Furuya, T, Ikura, M. | | Deposit date: | 1998-11-20 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase.
Nat.Struct.Biol., 6, 1999
|
|
1CKL
 
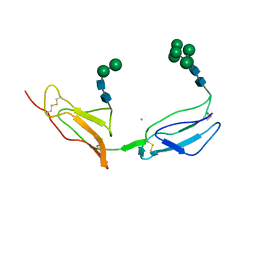 | | N-TERMINAL TWO DOMAINS OF HUMAN CD46 (MEMBRANE COFACTOR PROTEIN, MCP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Casasnovas, J, Larvie, M, Stehle, T. | | Deposit date: | 1999-04-22 | | Release date: | 1999-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of two CD46 domains reveals an extended measles virus-binding surface.
EMBO J., 18, 1999
|
|
1CKM
 
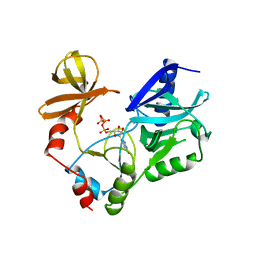 | |
1CKN
 
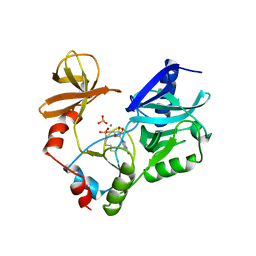 | | STRUCTURE OF GUANYLYLATED MRNA CAPPING ENZYME COMPLEXED WITH GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, MRNA CAPPING ENZYME, ... | | Authors: | Hakansson, K, Doherty, A.J, Wigley, D.B. | | Deposit date: | 1997-04-20 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes.
Cell(Cambridge,Mass.), 89, 1997
|
|
1CKO
 
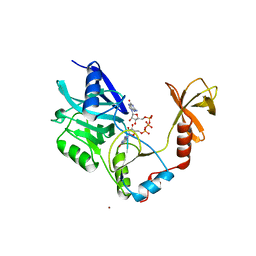 | |
1CKP
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR PURVALANOL B | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYCLIN-DEPENDENT PROTEIN KINASE 2), PURVALANOL B | | Authors: | Gray, N.S, Thunnissen, A.M.W.H, Schultz, P.G, Kim, S.H. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors.
Science, 281, 1998
|
|
1CKQ
 
 | |
1CKR
 
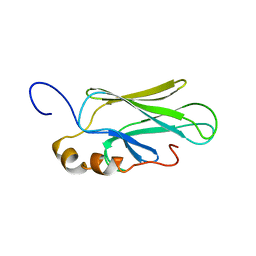 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
1CKS
 
 | |
1CKT
 
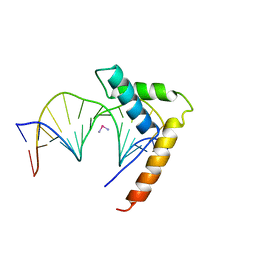 | | CRYSTAL STRUCTURE OF HMG1 DOMAIN A BOUND TO A CISPLATIN-MODIFIED DNA DUPLEX | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*(5IU)P*CP*TP*CP*TP*GP*GP*AP*CP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*GP*TP*CP*CP*AP*GP*AP*GP*AP*GP*G)-3'), ... | | Authors: | Ohndorf, U.-M, Rould, M.A, Pabo, C.O, Lippard, S.J. | | Deposit date: | 1999-04-23 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins.
Nature, 399, 1999
|
|
1CKU
 
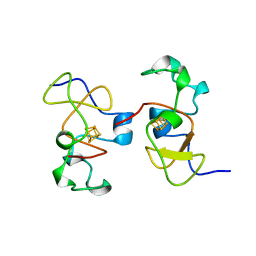 | | AB INITIO SOLUTION AND REFINEMENT OF TWO HIGH POTENTIAL IRON PROTEIN STRUCTURES AT ATOMIC RESOLUTION | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (HIPIP) | | Authors: | Parisini, E, Capozzi, F, Lubini, P, Lamzin, V, Luchinat, C, Sheldrick, G.M. | | Deposit date: | 1999-04-24 | | Release date: | 1999-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ab initio solution and refinement of two high-potential iron protein structures at atomic resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CKV
 
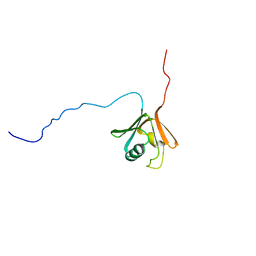 | |
1CKW
 
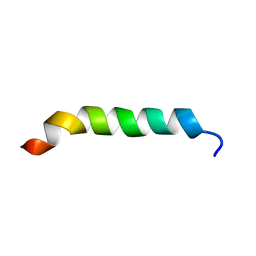 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKX
 
 | | Cystic fibrosis transmembrane conductance regulator: Solution structures of peptides based on the Phe508 region, the most common site of disease-causing Delta-F508 mutation | | Descriptor: | Cystic fibrosis transmembrane conductance regulator (CFTR) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKY
 
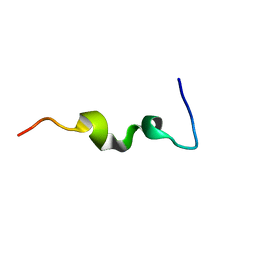 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKZ
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CL0
 
 | |
1CL1
 
 | |
