1JV9
 
 | | NMR Structure of BPTI Mutant G37A | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Battiste, J.L, Li, R, Woodward, C. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A highly destabilizing mutation, G37A, of the bovine pancreatic trypsin inhibitor retains the average native conformation but greatly increases local flexibility
Biochemistry, 41, 2002
|
|
1JV8
 
 | | NMR Structure of BPTI Mutant G37A | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Battiste, J.L, Li, R, Woodward, C. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A highly destabilizing mutation, G37A, of the bovine pancreatic trypsin inhibitor retains the average native conformation but greatly increases local flexibility
Biochemistry, 41, 2002
|
|
2N0O
 
 | |
1E8L
 
 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1B4O
 
 | |
2RRS
 
 | |
1B1V
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | Descriptor: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | Authors: | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | Deposit date: | 1998-11-23 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
1C7W
 
 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
1C7V
 
 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
1C9F
 
 | |
1BNO
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BZF
 
 | | NMR SOLUTION STRUCTURE AND DYNAMICS OF THE COMPLEX OF LACTOBACILLUS CASEI DIHYDROFOLATE REDUCTASE WITH THE NEW LIPOPHILIC ANTIFOLATE DRUG TRIMETREXATE, 22 STRUCTURES | | Descriptor: | DIHYDROFOLATE REDUCTASE, TRIMETREXATE | | Authors: | Polshakov, V.I, Birdsall, B, Frenkiel, T.A, Gargaro, A.R, Feeney, J. | | Deposit date: | 1998-10-28 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics in solution of the complex of Lactobacillus casei dihydrofolate reductase with the new lipophilic antifolate drug trimetrexate.
Protein Sci., 8, 1999
|
|
1BNP
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1B5N
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | Descriptor: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | Authors: | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
1CQL
 
 | |
1EKA
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*GP*CP*UP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1HLS
 
 | | NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) | | Descriptor: | INSULIN | | Authors: | Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1995-06-28 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an engineered biologically potent insulin monomer, B16 Tyr-->His, as determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1I93
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND D16 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND D16 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-17 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1KQI
 
 | | NMR Solution Structure of the trans Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
1I98
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND D18 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND D18 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-18 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1PEH
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPNH1 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
1PEI
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPC22 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
1PES
 
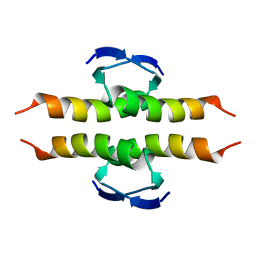 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
