6CEJ
 
 | |
1AH8
 
 | |
1AH6
 
 | |
4QJV
 
 | |
4QIW
 
 | |
6TNI
 
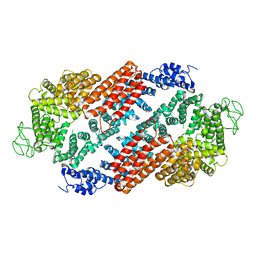 | | Structure of FANCD2 homodimer | | Descriptor: | Uncharacterized protein | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5WFY
 
 | |
6JGR
 
 | | Crystal structure of barley exohydrolaseI W434Y in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6VAA
 
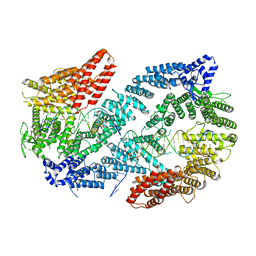 | | Structure of the Fanconi Anemia ID complex bound to ICL DNA | | Descriptor: | DNA (26-MER), DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex.
Nature, 580, 2020
|
|
6JG6
 
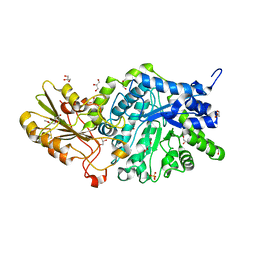 | | Crystal structure of barley exohydrolaseI W286A mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6VAD
 
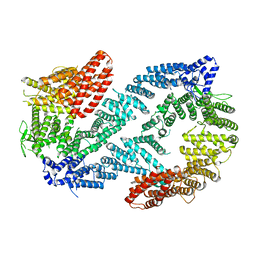 | | Fanconi Anemia ID complex | | Descriptor: | Fanconi anemia group D2 protein, Fanconi anemia, complementation group I | | Authors: | Pavletich, N.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex.
Nature, 580, 2020
|
|
6JGT
 
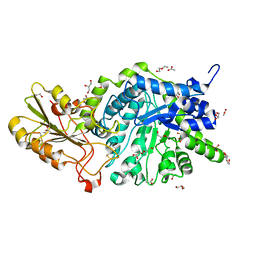 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JG1
 
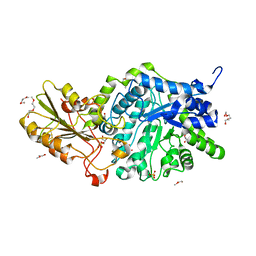 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGG
 
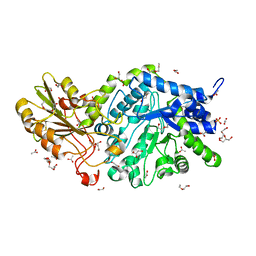 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGB
 
 | | Crystal structure of barley exohydrolaseI W286F mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGQ
 
 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGE
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JG7
 
 | | Crystal structure of barley exohydrolaseI W286F in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGN
 
 | | Crystal structure of barley exohydrolaseI W434H in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGC
 
 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGS
 
 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGK
 
 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JG2
 
 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-nitrophenoxy)-3,5-bis(oxidanyl)oxan-4-yl]sulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGD
 
 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGP
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
