5LZ9
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 2-fluoranyl-5-[2-[(4~{S})-4-[4-methyl-1,1-bis(oxidanylidene)thian-4-yl]-2-oxidanylidene-pyrrolidin-1-yl]ethoxy]benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase, ... | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
7AHS
 
 | | titin-N2A Ig81-Ig83 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Isoform 5 of Titin, ... | | Authors: | Fleming, J.R, Mayans, O. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The N2A region of titin has a unique structural configuration.
J.Gen.Physiol., 153, 2021
|
|
3ZUC
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus determined from the crystals grown in the presence of Nickel | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-18 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5VRI
 
 | | Crystal structure of SsoPox AsA6 mutant (F46L-C258A-W263M-I280T) - closed form | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
8K3J
 
 | | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2, ... | | Authors: | Zhang, Z.Z. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer
To Be Published
|
|
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
3MBS
 
 | | Crystal structure of 8mer PNA | | Descriptor: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
5F4H
 
 | | Archael RuvB-like Holiday junction helicase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleotide binding protein PINc | | Authors: | Zhai, B, DuPrez, K.T, Doukov, T.I, Shen, Y, Fan, L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.
J. Mol. Biol., 429, 2017
|
|
4NRB
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-1 N01197 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Authors: | Muniz, J.R.C, Felletar, I, Chaikuad, A, Filippakopoulos, P, Ferguson, F.M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ciulli, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
6EO6
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1H-indol-3-yl)acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA63A - TBA MODIFIED APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6JAL
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in ligand free form | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6CZA
 
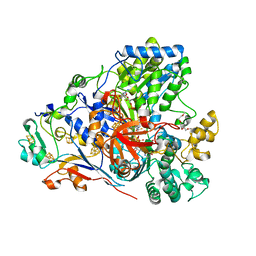 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 bound to phosphate | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | Authors: | Glasser, N.R, Newman, D.K. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3HM4
 
 | |
5LGS
 
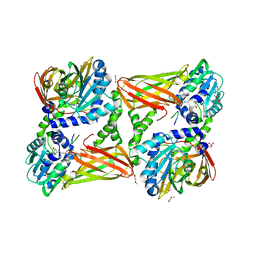 | | Crystal structure of mouse CARM1 in complex with ligand P2C3u | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7V0Q
 
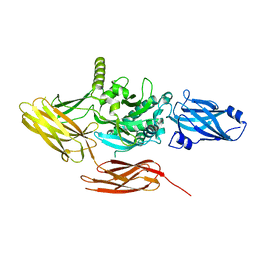 | | Local refinement of protein 4.2, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | Protein 4.2 | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-10 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UZQ
 
 | | Local refinement of RhAG-RhCE-ANK1(AR1-5), from consensus refinement of all classes | | Descriptor: | Ammonium transporter Rh type A, Ankyrin-1, Blood group Rh(CE) polypeptide, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5D5U
 
 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4NR2
 
 | | Crystal structure of STK4 (MST1) SARAH domain | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 4 | | Authors: | Chaikuad, A, Krojer, T, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of STK4 (MST1) SARAH domain
To be Published
|
|
7AOT
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6M7I
 
 | | Crystal structure of KPC-2 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
4HVD
 
 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
3HKR
 
 | | Crystal Structure of Glutathione Transferase Pi Y108V Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
6MA2
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor ent-1a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2S)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
5D2K
 
 | |
7VRX
 
 | | Pad-1 in the absence of substrate | | Descriptor: | Aminotransferase, SULFATE ION | | Authors: | Choi, M, Rhee, S. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96634674 Å) | | Cite: | Structural and biochemical basis for the substrate specificity of Pad-1, an indole-3-pyruvic acid aminotransferase in auxin homeostasis.
J.Struct.Biol., 214, 2022
|
|
