1IAB
 
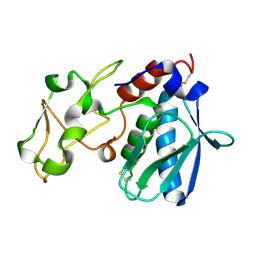 | | CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY | | Descriptor: | ASTACIN, COBALT (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures, spectroscopic features, and catalytic properties of cobalt(II), copper(II), nickel(II), and mercury(II) derivatives of the zinc endopeptidase astacin. A correlation of structure and proteolytic activity.
J.Biol.Chem., 269, 1994
|
|
1OCT
 
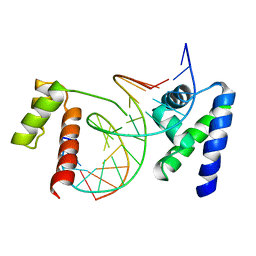 | | CRYSTAL STRUCTURE OF THE OCT-1 POU DOMAIN BOUND TO AN OCTAMER SITE: DNA RECOGNITION WITH TETHERED DNA-BINDING MODULES | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), PROTEIN (OCT-1 POU DOMAIN) | | Authors: | Klemm, J.D, Rould, M.A, Aurora, R, Herr, W, Pabo, C.O. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules.
Cell(Cambridge,Mass.), 77, 1994
|
|
1BTA
 
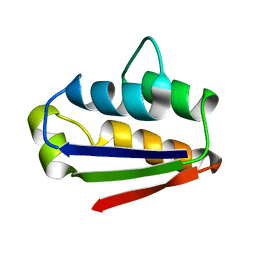 | |
1IAD
 
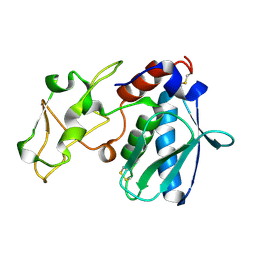 | | REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON TO THERMOLYSIN | | Descriptor: | ASTACIN | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.
J.Mol.Biol., 229, 1993
|
|
1BTB
 
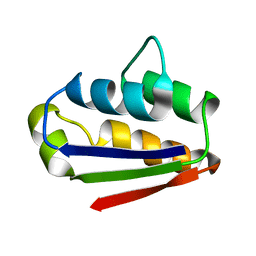 | |
1TNR
 
 | |
1IAC
 
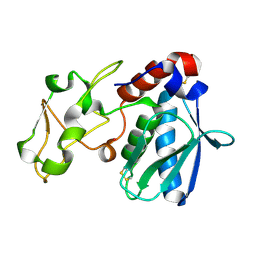 | | REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON WITH THERMOLYSIN | | Descriptor: | ASTACIN, MERCURY (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.
J.Mol.Biol., 229, 1993
|
|
1IAA
 
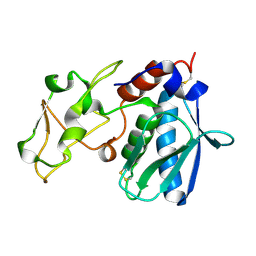 | | CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY | | Descriptor: | ASTACIN, COPPER (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, spectroscopic features, and catalytic properties of cobalt(II), copper(II), nickel(II), and mercury(II) derivatives of the zinc endopeptidase astacin. A correlation of structure and proteolytic activity.
J.Biol.Chem., 269, 1994
|
|
1IAE
 
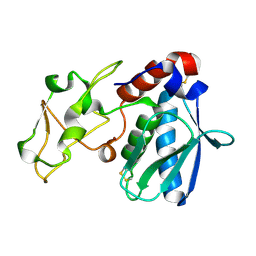 | | CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY | | Descriptor: | ASTACIN, NICKEL (II) ION | | Authors: | Grams, F, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures, spectroscopic features, and catalytic properties of cobalt(II), copper(II), nickel(II), and mercury(II) derivatives of the zinc endopeptidase astacin. A correlation of structure and proteolytic activity.
J.Biol.Chem., 269, 1994
|
|
1IAG
 
 | |
1AIA
 
 | |
1AIC
 
 | |
1AIB
 
 | |
1TRU
 
 | |
1TRW
 
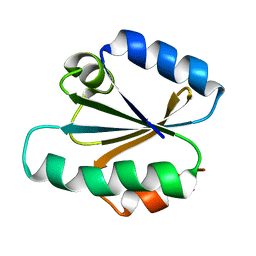 | |
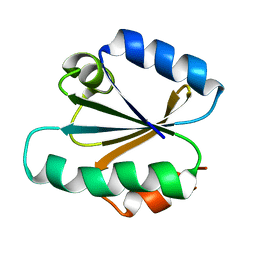
1TRV
 
 | |
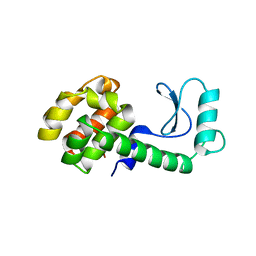
216L
 
 | |
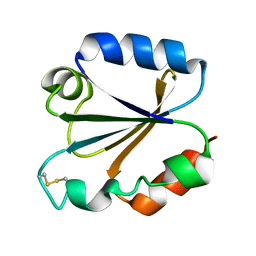
1TRS
 
 | |
1COE
 
 | |
1COD
 
 | |
1LEA
 
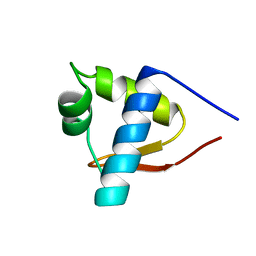 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
1LEB
 
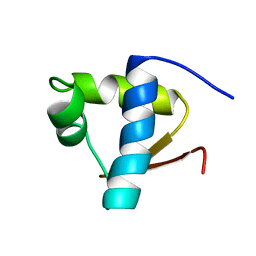 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
1HMA
 
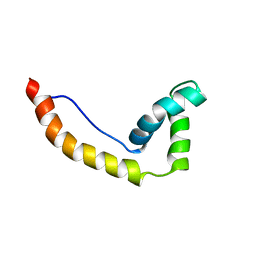 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
1AGM
 
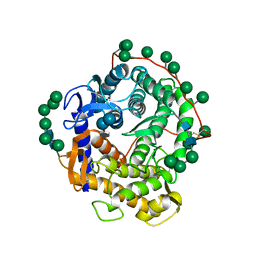 | | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-05-13 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
J.Biol.Chem., 269, 1994
|
|
1LNE
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CADMIUM ION, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
