1D6G
 
 | |
4A5V
 
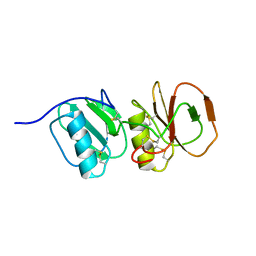 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
1J79
 
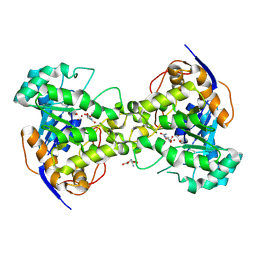 | | Molecular Structure of Dihydroorotase: A Paradigm for Catalysis Through the Use of a Binuclear Metal Center | | Descriptor: | N-CARBAMOYL-L-ASPARTATE, OROTIC ACID, ZINC ION, ... | | Authors: | Thoden, J.B, Phillips Jr, G.N, Neal, T.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center.
Biochemistry, 40, 2001
|
|
6HHW
 
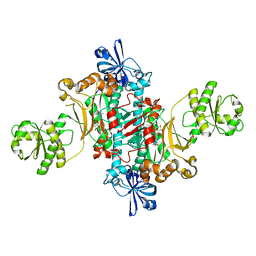 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)uridine | | Descriptor: | 5'-O-(N-(L-aspartyl)-sulfamoyl)uridine, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
5K50
 
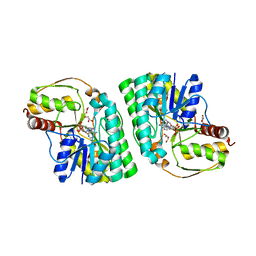 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ and L-allo-threonine refined to 2.23 angstroms | | Descriptor: | ACETATE ION, ALLO-THREONINE, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6R2U
 
 | | Zinc-alpha2-Glycoprotein with a Fluorescent Dansyl C 11 Fatty Acid | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, AZIDE ION, SULFATE ION, ... | | Authors: | Lau, A.M, Gor, J, Perkins, S.J, Coker, A.R, McDermott, L.C. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of zinc-alpha 2-glycoprotein in complex with a fatty acid reveals multiple different modes of protein-lipid binding.
Biochem.J., 476, 2019
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6ZSC
 
 | | Human mitochondrial ribosome in complex with E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
4B2J
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Staudt, H, Hoesl, M, Dreuw, A, Serdjukow, S, Oesterhelt, D, Budisa, N, Wachtveitl, J, Grininger, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed Manipulation of a Flavoprotein Photocycle.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4ZRC
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3FSB
 
 | | Crystal structure of QdtC, the dTDP-3-amino-3,6-dideoxy-D-glucose N-acetyl transferase from Thermoanaerobacterium thermosaccharolyticum in complex with CoA and dTDP-3-amino-quinovose | | Descriptor: | COENZYME A, QdtC, [(3R,4S,5S,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Studies of QdtC: an N-Acetyltransferase Required for the Biosynthesis of dTDP-3-Acetamido-3,6-Dideoxy-alpha-D-Glucose.
Biochemistry, 48, 2009
|
|
1LFH
 
 | | MOLECULAR REPLACEMENT SOLUTION OF THE STRUCTURE OF APOLACTOFERRIN, A PROTEIN DISPLAYING LARGE-SCALE CONFORMATIONAL CHANGE | | Descriptor: | CHLORIDE ION, LACTOFERRIN | | Authors: | Anderson, B.F, Baker, E.N, Norris, G.E. | | Deposit date: | 1991-09-04 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular replacement solution of the structure of apolactoferrin, a protein displaying large-scale conformational change.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
3FSC
 
 | | Crystal structure of QdtC, the dTDP-3-amino-3,6-dideoxy-D-glucose N-acetyl transferase from Thermoanaerobacterium thermosaccharolyticum in complex with CoA and dTDP-3-amino-fucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, COENZYME A, QdtC | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of QdtC: an N-Acetyltransferase Required for the Biosynthesis of dTDP-3-Acetamido-3,6-Dideoxy-alpha-D-Glucose.
Biochemistry, 48, 2009
|
|
3FS8
 
 | | Crystal structure of QdtC, the dTDP-3-amino-3,6-dideoxy-D-glucose N-acetyl transferase from Thermoanaerobacterium thermosaccharolyticum in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, QdtC, THYMINE | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of QdtC: an N-Acetyltransferase Required for the Biosynthesis of dTDP-3-Acetamido-3,6-Dideoxy-alpha-D-Glucose.
Biochemistry, 48, 2009
|
|
6GTJ
 
 | | Neutron crystal structure for copper nitrite reductase from Achromobacter Cycloclastes at 1.8 A resolution | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Antonyuk, S.V, Blakeley, M.P, Halsted, T.P, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | NEUTRON DIFFRACTION (1.801 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
4V73
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre5a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4V
 
 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4X
 
 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7W4U
 
 | |
4BCC
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND P2- substituted N-acyl-prolylpyrrolidine inhibitor | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | VanDerVeken, P, Fulop, V, Rea, D, Gerard, M, VanElzen, R, Joossens, J, Cheng, J.D, Baekelandt, V, DeMeester, I, Lambeir, A.M, Augustyns, K. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy.
J.Med.Chem., 55, 2012
|
|
7W4S
 
 | |
7W4V
 
 | |
4H5V
 
 | | HSC70 NBD with Mg | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
